Protein-Carbohydrate Interactions as Part of Plant Defense and Animal Immunity
Abstract
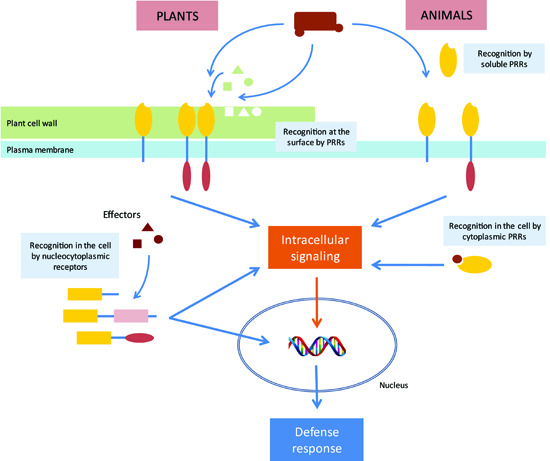
:1. Introduction
| Plant Lectin Family | Typical Saccharide Ligands | Predicted Localization |
| Agaricus bisporus lectin family | GlcNAc/GalNAc, Galactose | Nucleus, cytosol |
| Amaranthin family | GalNAc | Nucleus, cytosol |
| Chitinase related agglutinin family | High mannose N-glycans | Vacuole, membrane bound |
| Cyanovirin family | Mannose | Nucleus |
| Euonymus europaeus lectin family | Galactosides, high-mannose N-glycans | Nucleus, cytosol |
| Galanthus nivalis lectin family | Mannose | Vacuole, nucleus, cytosol or membrane bound |
| Hevein family | Chitin | Vacuole |
| Jacalin family | Mannose- and galactose-specific subgroup | Nucleus, cytosol, vacuole |
| Legume family | Mannose | Vacuole, nucleus, cytosol or membrane bound |
| LysM family | Chitin, peptidoglycan | Vacuole, nucleus, cytosol or membrane bound |
| Nicotiana tabacum lectin family | (GlcNAc)n, high-mannose and complex N-glycans | Nucleus, cytosol |
| Ricin-B family | Gal/GalNAc, Sialylated Gal/GalNAc | Vacuole, nucleus, cytosol |
| Animal Lectin Family * | Typical Saccharide Ligands | Predicted Localization |
| Calnexin and Calreticulin | Glc1Man9 | ER |
| M-type lectins | Man8 | ER |
| L-type lectins | Various | ER, Golgi |
| P-type lectins | Man6-phosphate | Secretory pathway |
| C-type lectins | Mannosides, galactosides, sialic acids and others | Membrane bound, extracellular |
| S-type lectins (galectins) | β-galactosides | Cytosol, extracellular |
| I-type lectins (siglecs) | Sialic acid | Membrane bound |
| R-type lectins | Various | Golgi, membrane bound |
| F-box lectins | GlcNAc2 of N-glycans | Cytoplasma |
| Fibrinogen-type lectin | GlcNAc, GalNAc | Membrane bound, extracellular |
| Chi-lectins | Chito-oligosaccharides | Extracellular |
| F-type lectins | Fucose terminating oligosaccharides | Extracellular |
| Intelectins | Galactose, galactofuranose, pentoses | Membrane bound, extracellular |
| Annexins | Glycosaminoglycans, heparin and heparin sulfate | Membrane bound |
2. Lectins as Pattern Recognition Receptors in the Innate Immune System
2.1. Animal Lectin PRRs

2.2. Plant Lectin PRRs

2.3. Plant Effector Triggered Immunity
3. Autophagy
4. Non-PRR Function of Lectins in the Immune Response
5. Adaptive Immunity and Epigenetic Imprinting
5.1. Adaptive Immunity
5.2. Epigenetic Imprinting
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boyd, W.C.; Shapleigh, E. Specific precipitating activity of plant agglutinins (Lectins). Science 1954, 119, 419. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Lannoo, N.; Peumans, W.J. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar]
- Drickamer, K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J. Biol. Chem. 1988, 263, 9557–9560. [Google Scholar] [PubMed]
- Kilpatrick, D.C. Animal lectins: A historical introduction and overview. Biochim. Biophys. Acta 2002, 1572, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.S. Lectins: An overview. In Animal Lectins: Form, Function and Clinical Applications; Springer-Verlag: Wien, Austria, 2012; pp. 3–25. [Google Scholar]
- Van Damme, E.J.M.; Barre, A.; Rougé, P.; Peumans, W.J. Cytoplasmic/nuclear plant lectins: A new story. Trends Plant Sci. 2004, 9, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Lannoo, N.; Fouquaert, E.; Peumans, W.J. The identification of inducible cytoplasmic/nuclear carbohydrate-binding proteins urges to develop novel concepts about the role of plant lectins. Glycoconj. J. 2004, 20, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, B.; Ramar, M. Detection and characterization of natural and inducible lectins in human serum. Res. Immunol. 2012, 2, 132–141. [Google Scholar] [CrossRef]
- Johnson, R.Q.; Lindsay, R.J.; Petridis, L.; Shen, T. Investigation of carbohydrate recognition via computational simulation. Molecules 2015, 20, 7700–7718. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K. Introduction to Lectin Families. Available online: https://www.imperial.ac.uk/animallectins/ctld/lectins.html (accessed on 2 February 2015).
- Drickamer, K. Making a fitting choice: Common aspects of sugar-binding sites in plant and animal lectins. Structure 1997, 5, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.E.; Drickamer, K. Convergent and divergent mechanisms of sugar recognition across kingdoms. Curr. Opin. Struct. Biol. 2014, 28, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Loris, R. Principles of structures of animal and plant lectins. Biochim. Biophys. Acta 2002, 1572, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Delaloye, J.; Calandra, T. Host innate immune responses to microbial pathogens. Curr. Vasc. Pharmacol. 2013, 11, 123–132. [Google Scholar] [PubMed]
- Ni, Y.; Tizard, I. Lectin-carbohydrate interaction in the immune system. Vet. Immunol. Immunopathol. 1996, 55, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Che, C.Y.; Zhang, J.F.; Lee, J.E.; Lin, J.; Hu, L.T.; Jiang, N.; Wang, Q.; Xu, Q.; Zhao, G.Q. Early expression of mannose-binding lectin 2 during Aspergillus fumigatus infection in human corneal epithelial cells. Int. J. Ophthalmol. 2015, 8, 35–38. [Google Scholar] [PubMed]
- Holmskov, U.; Malhotra, R.; Sim, R.B.; Jensenius, J.C. Collectins: Collagenous C-type lectins of the innate immune defense system. Immunol. Today 1994, 15, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.H.; Thiel, S.; Widemann, H.; Timpl, R.; Reid, K.B.M. Binding of pentamer/hexamer forms of mannan-binding protein to zymosan activates the proenzyme c1r2cls2 complex, of the classical pathway of complement, without involvement of C1q. J. Immunol. 1990, 144, 2287–2294. [Google Scholar] [PubMed]
- Steel, D.M.; Whitehead, A.S. The major acute phase reactants: C-reactive protein, serum amyloid P component and serum amyloid A protein. Immunol. Today 1994, 15, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Gewurz, H.; Zhang, X.H.; Lint, T.F. Structure and function of the pentraxins. Curr. Opin. Immunol. 1995, 7, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.D.; Ezekowitz, R.A. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. 1998, 10, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Wileman, T.E.; Lennartz, M.R.; Stahl, P.D. Identification of the macrophage mannose receptor as a 175-kDa membrane protein. Proc. Natl. Acad. Sci. USA 1986, 83, 2501–2505. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.E.; Bezouska, K.; Drickamer, K. Contribution to ligand binding by multiple carbohydrate-recognition domains in the macrophage mannose receptor. J. Biol. Chem. 1992, 267, 719–726. [Google Scholar] [PubMed]
- Taylor, M.E.; Drickamer, K. Structural requirements for high affinity binding of complex ligands by the macrophage mannose receptor. J. Biol. Chem. 1993, 268, 399–404. [Google Scholar] [PubMed]
- Goren, M.B.; D’Arcy Hart, P.; Young, M.R.; Armstrong, J.A. Prevention of phagosome-lysosome fusion in cultured macrophages by sulfatides of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1976, 73, 2510–2514. [Google Scholar] [CrossRef] [PubMed]
- Benoff, S.; Cooper, G.W.; Hurley, I.; Napolitano, B.; Rosenfeld, D.L.; Scholl, G.M.; Hershlag, A. Human sperm fertilizing potential in vitro is correlated with differential expression of a head-specific mannose-ligand receptor. Fertil. Steril. 1993, 59, 854–862. [Google Scholar] [PubMed]
- Leteux, C.; Chai, W.; Loveless, R.W.; Yuen, C.T.; Uhlin-Hansen, L.; Combarnous, Y.; Jankovic, M.; Maric, S.C.; Misulovin, Z.; Nussenzweig, M.C.; et al. The cysteine-rich domain of the macrophage mannose receptor is a multispecific lectin that recognizes chondroitin sulfates A and B and sulfated oligosaccharides of blood group Lewisa and Lewisx types in addition to the sulfated N-glycans of lutropin. J. Exp. Med. 2000, 191, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.; Six, H.; Rodman, J.S.; Schlesinger, P.; Tulsiani, D.R.; Touster, O. Evidence for specific recognition sites mediating clearance of lysosomal enzymes in vivo. Proc. Natl. Acad. Sci. USA 1976, 73, 4045–4049. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Evers, S.; Roeder, D.; Parlow, A.F.; Risteli, J.; Risteli, L.; Lee, Y.C.; Feizi, T.; Langen, H.; Nussenzweig, M.C. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science 2002, 295, 1898–1901. [Google Scholar] [CrossRef] [PubMed]
- Gantner, B.N.; Simmons, R.M.; Canavera, S.J.; Akira, S.; Underhill, D.M. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003, 197, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D. Dectin-1: A signalling non-TLR pattern-recognition receptor. Nat. Rev. Immunol. 2006, 6, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Brunner, F. Innate immunity in plants and animals: Emerging parallels between recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Brunner, F.; Kemmerlinck, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V.; Roux, M.; Zipfel, C. Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009, 150, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Wirthmueller, L.; Maqbool, A.; Banfield, M.J. On the front line: Structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 2013, 11, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J.M. Lectin domains at the frontiers of plant defense. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Zimmerli, L. Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Sherman-Broyles, S.; Boggs, N.; Farkas, A.; Liu, P.; Vrebalov, J.; Nasrallah, M.E.; Nasrallah, J.B. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 2007, 19, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Govers, F. Arabidopsis L-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J. Exp. Bot. 2009, 60, 4383–4396. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.T.; Oh, J.; Kim, K.H.; Uhm, J.Y.; Lee, B.M. Isolation and characterization of NgRLK1 a receptor-like kinase of Nicotiana glutinosa that interacts with the elicitin of Phytophthora capsici. Mol. Biol. Rep. 2009, 37, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.C.; Mishra, S.; Tsai, C.H.; Chien, C.C.; Chen, C.W.; Desclos-Theveniau, M.; Chu, P.W.; Schulze, B.; Chinchilla, D.; et al. The lectin-receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 2012, 24, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Desclos-Theveniau, M.; Arnaud, D.; Huang, T.Y.; Lin, G.J.; Chen, W.Y.; Lin, Y.C.; Zimmerli, L. The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2012, 8, e1002513. [Google Scholar] [CrossRef]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Cambi, A.; Koopman, M.; Figdor, C.G. How C-type lectins detect pathogens. Cell Microbiol. 2005, 7, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.K.; Tuteja, N. Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 2012, 80, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; de Sain, M.; Weide, R.; Gouget, A.; Klamer, S.; Canut, H.; Govers, F. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011, 7, e1001327. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ju, H.W.; Min, J.H.; Zhang, X.; Kim, S.H.; Yang, K.Y.; Kim, C.S. Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 2013, 203–204, 98–106. [Google Scholar] [PubMed]
- Huang, P.Y.; Yeh, Y.H.; Liu, A.C.; Cheng, C.P.; Zimmerli, L. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014, 79, 243–255. [Google Scholar]
- Cao, Y.; Tanaka, K.; Nguyen, C.T.; Stacey, G. Extracellular ATP is a central signaling molecule in plant stress responses. Curr. Opin. Plant Biol. 2014, 20, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacy, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Gronlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Knogge, W.; Scheel, D. LysM receptors recognize friend and foe. Proc. Natl. Acad. Sci. USA 2006, 103, 10829–10830. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.R.; Fisher, E.J.; Chang, J.H.; Mole, B.M.; Dangl, J.L. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 2006, 60, 425–449. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Toruño, T.Y.; Elowsky, C.G.; Zhang, C.; Steinbrenner, J.; Beynon, J.; Alfano, J.R. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2013, 201, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nurnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Mehra, A.; Zahra, A.; Thompson, V.; Sirisaengtaksin, N.; Wells, A.; Porto, M.; Köster, S.; Penberthy, K.; Kubota, Y.; Dricot, A.; et al. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog. 2013, 9, e1003734. [Google Scholar] [CrossRef] [PubMed]
- McCollister, B.D.; Vazquez-Torres, A. Interactions of Salmonella enterica with phagocytic cells. In Salmonella Infections Clinical, Immunological and Molecular Aspects; Mastroeni, P., Maskell, D., Eds.; Cambridge University Press: Cambridge, UK, 2006; pp. 225–278. [Google Scholar]
- Huang, J.; Brumell, J.H. A Sweet way of sensing danger. Nature 2006, 482, 316–317. [Google Scholar] [CrossRef]
- Birmingham, C.L.; Smith, A.C.; Bakowski, M.A.; Yoshimori, T.; Brumell, J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006, 281, 11374–11383. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, S.; Omori, H.; Saitoh, T.; Sone, T.; Guan, J.L.; Akira, S.; Imamoto, F.; Noda, T.; Yoshimori, T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell 2011, 22, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Blommaart, E.; Luiken, J.; Meijer, A. Autophagic proteolysis: Control and specificity. Histochem. J. 1997, 29, 365–385. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Thurston, T.L.M.; Wandel, M.P.; von Muhlinen, N.; Foeglein, A.; Randow, F. Galectin-8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012, 482, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.; Jiang, X.; Birmingham, C.; So, N.; Brumell, J. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Current Biology 2004, 14, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of intracellular Shigella from autophagy. Science 2005, 307, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis. Apoptosis 2014, 19, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Wolf, M.Y.; Wolf, Y.I.; Koonin, E.V. The origins of phagocytosis and eukaryogenesis. Biol. Direct 2009, 4, 9:1–9:26. [Google Scholar]
- Brewin, N.J. Tissue and cell invasion by Rhizobium: The structure and development of infection threads and symbiosisomes. In The Rhizobiaceae; Spaink, H.P., Kondorosi, A., Hooykaas, P.J.J., Eds.; Kluwer academic: Dordrecht, The Netherlands, 1998; pp. 417–429. [Google Scholar]
- Parniske, M. Intracellular accommodation of microbes by plants: A common developmental program for symbiosis and disease. Curr. Opin. Plant Biol. 2000, 3, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.J.; Fukao, Y.; Fujiwara, M.; Nishimura, M.; Hara-Nishimura, I. Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell. Physiol. 2008, 49, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Hayashi, Y.; Yamada, K.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell. Physiol. 2003, 44, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Honig, A.; Levanony, H.; Peled-Zehavi, H.; Galili, G. Arabidopsis ATG8-INTERACTING PROTEIN1 Is Involved in Autophagy-Dependent Vesicular Trafficking of Plastid Proteins to the Vacuole. Plant Cell 2014, 26, 4084–4101. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [PubMed]
- Levine, B.; Yuan, J. Autophagy in cell death: An innocent convict? J. Clin. Investig. 2005, 115, 2679–2688. [Google Scholar] [CrossRef] [PubMed]
- Seay, M.; Dinesh-Kumar, S.P.; Levine, B. Digesting oneself and digesting microbes: Autophagy as a host response to viral infection. In Modulation of Host Gene Expression and Innate Immunity by Viruses; Palese, P., Ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 245–279. [Google Scholar]
- Seay, M.; Patel, S.; Dinesh-Kumar, S.P. Autophagy and plant innate immunity. Cell. Microbiol. 2006, 8, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Selectin ligands. Proc. Nat. Acad. Sci. USA 1994, 91, 7390–7397. [Google Scholar] [CrossRef] [PubMed]
- Muerkoster, S.; Rocha, M.; Crocker, P.R.; Schirrmacher, V.; Umanski, V. Sialoadhesin-positive host macrophages play an essential role in graft-versus-leukemia reactivity in mice. Blood 1999, 93, 4375–4386. [Google Scholar] [PubMed]
- Crocker, P.R.; Varki, A. Siglecs in the immune system. Immunology 2001, 103, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Lanier, L.L. Immune inhibitory receptors. Science 2000, 290, 84–89. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, T.L.; Williams, G.T.; Batista, F.D.; Neuberger, M.S. Deficiency in CD22, a B cell-specific inhibitory receptor, is sufficient to predispose to development of high affinity antibodies. J. Exp. Med. 1999, 189, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Fouquaert, E.; Peumans, W.J.; Vandekerckhove, T.; Ongenaert, M.; Van Damme, E.J.M. Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol. 2009, 9, 136:1–136:17. [Google Scholar] [CrossRef] [Green Version]
- Fouquaert, E.; Van Damme, E.J.M. Promiscuity of the Euonymus carbohydrate-binding domain. Biomolecules 2012, 2, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, X.; Yuan, D.; Zhang, L.; Jiang, X.; Tao, Z.; Li, Y.; Wang, J.; Li, X.; Yang, Y. Over-expression of ArathEULS3 confers ABA sensitivity and drought tolerance in Arabidopsis. Plant Cell Tissue Organ Cult. 2014, 117, 431–442. [Google Scholar] [CrossRef]
- Berendzen, K.W.; Böhmer, M.; Wallmeroth, N.; Peter, S.; Vesić, M.; Zhou, Y.; Tiesler, F.K.; Schleifenbaum, F.; Harter, K. Screening for in planta protein–protein interactions combining bimolecular fluorescence complementation with flow cytometry. Plant Methods 2012, 8, 25:1–25:17. [Google Scholar] [CrossRef]
- Bourne, Y.; Astoul, C.H.; Zamboni, V.; Peumans, W.J.; Menu-Bouaouiche, L.; Van Damme, E.J.M.; Barre, A.; Rougé, P. Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose. Biochem. J. 2002, 364, 173–180. [Google Scholar] [PubMed]
- Peumans, W.J.; Hause, B.; Van Damme, E.J.M. The galactose-binding and mannose-binding jacalin-related lectins are located in different sub-cellular compartments. FEBS Lett. 2000, 477, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Xu, W.; Xiang, Y.; Jia, H.; Zhang, L.; Ma, Z. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.M.; Zhang, Q.; Zhao, W.S.; Wang, Y.Y.; Peng, Y.L. Identification of a lectin gene induced in rice in response to Magnaporthe grisea infection. Acta Bot. Sin. 2003, 23, 799–810. [Google Scholar]
- De Souza Filho, G.A.; Ferreira, B.S.; Dias, J.M R.; Queiroz, K.S.; Branco, A.T.; Bressan-Smith, R.E.; Oliveira, J.G.; Garcia, A.B. Accumulation of SALT protein in rice plants as a response to environmental stresses. Plant Sci. 2003, 164, 623–628. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, M.; Wei, Z.; Tong, J.; Zhang, L.; Xiao, L.; Ma, Z.; Wang, Y. A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 2011, 62, 5471–5483. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, Y.; Maejima, K.; Ozeki, J.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; et al. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 2012, 24, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Regente, M.; Taveira, G.B.; Pinedo, M.; Elizalde, M.M.; Ticchi, A.J.; Diz, M.S.S.; Carvalho, A.O.; de la Canal, L.; Gomes, V.M. A sunflower lectin with antifungal properties and putative medical mycology applications. Curr. Microbiol. 2014, 69, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Kanagawa, M.; Liu, Y.; Hanashima, S.; Ikeda, A.; Chai, W.; Nakano, Y.; Kojima-Aikawa, K.; Feizi, T.; Yamaguchi, Y. Structural basis for multiple sugar recognition of Jacalin-related human ZG16p lectin. J. Biol. Chem. 2014, 289, 16954–16965. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Lee, J.; Lee, J. Community-based network study of protein-carbohydrate interactions in plant lectins using glycan array data. PLoS ONE 2014, 9, e95480. [Google Scholar] [CrossRef] [PubMed]
- Beneteau, J.; Renard, D.; Marché, L.; Douville, E.; Lavenant, L.; Rahbé, Y.; Dupont, D.; Vilaine, F.; Dinant, S. Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 2010, 153, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- R-type lectins. In Essentials of Glycobiology, 2nd ed.; Varki, E.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. (Eds.) Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006; pp. 403–414.
- Shang, C.; Van Damme, E.J.M. Comparative analysis of carbohydrate binding properties of Sambucus nigra lectins and ribosome-inactivating proteins. Glycoconj. J. 2014, 31, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Kerringan, A.M.; Brown, G.D. Syk-coupled C-type lectins in immunity. Trends Immunol. 2011, 32, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 2010, 327, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Systemic acquired resistance. Plant Signal. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful ‘‘memories’’ of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Beckers, G.J.M.; Jaskiewicz, M.; Liu, Y.; Underwood, W.R.; He, S.Y.; Zhang, S.; Conrath, U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 2009, 21, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Amasino, R.M. Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 2004, 7, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Xu, S.; Li, C.; Xu, Y.; Xing, L.; Niu, Y.; Huan, Q.; Tang, Y.; Zhao, C.; Wagner, D.; et al. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 2014, 5, 4572:1–4572:13. [Google Scholar] [CrossRef]
- Chen, Y.; Peumans, W.; Hause, B.; Bras, J.; Kumar, M.; Proost, P.; Barre, A.; Rougé, P.; Van Damme, E.J.M. Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chito-oligosaccharide binding lectin in tobacco leaves. FASEB J. 2002, 16, 905–907. [Google Scholar] [PubMed]
- Lannoo, N.; Vandenborre, G.; Miersch, O.; Smagghe, G.; Wasternack, C.; Peumans, W.J.; Van Damme, E.J.M. The jasmonate-induced expression of the Nicotiana tabacum leaf lectin. Plant Cell Physiol. 2007, 48, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Vandenborre, G.; Van Damme, E.J.M.; Smagghe, G. Natural products: Plant lectins as important tools in controlling pest insects. In Biorational Control of Arthropod Pests; Ishaaya, I., Horowitz, A.R., Eds.; Springer Science: Dordrecht, The Netherlands, 2009; pp. 163–187. [Google Scholar]
- Schouppe, D.; Ghesquière, B.; Menschaert, G.; De Vos, W.H.; Bourque, S.; Trooskens, G.; Proost, P.; Gevaert, K.; Van Damme, E.J.M. Interaction of the tobacco lectin with histone proteins. Plant Physiol. 2011, 155, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Delporte, A.; De Vos, W.H.; Van Damme, E.J.M. In vivo interaction between the tobacco lectin and the core histone proteins. J. Plant Physiol. 2014, 171, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Delporte, A.; Van Holle, S.; Lannoo, N.; Van Damme, E.J.M. The tobacco lectin, prototype of the family of Nictaba-related proteins. Curr. Prot. Pept. Sci. 2015, 16, 5–16. [Google Scholar] [CrossRef]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Schutter, K.; Van Damme, E.J.M. Protein-Carbohydrate Interactions as Part of Plant Defense and Animal Immunity. Molecules 2015, 20, 9029-9053. https://doi.org/10.3390/molecules20059029
De Schutter K, Van Damme EJM. Protein-Carbohydrate Interactions as Part of Plant Defense and Animal Immunity. Molecules. 2015; 20(5):9029-9053. https://doi.org/10.3390/molecules20059029
Chicago/Turabian StyleDe Schutter, Kristof, and Els J. M. Van Damme. 2015. "Protein-Carbohydrate Interactions as Part of Plant Defense and Animal Immunity" Molecules 20, no. 5: 9029-9053. https://doi.org/10.3390/molecules20059029