A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative
Abstract
:1. Introduction

2. Results and Discussion
2.1. UV-Vis Absorption Titration

| DNAs | 1 | 2 | Tm/°C | ∆Tm/°C | |||
|---|---|---|---|---|---|---|---|
| 10−6 K/M−1 | n | 10−6 K/M−1 | n | DNAs | 1 | 2 | |
| a-core (K+) | 10 ± 0.5 a | 1.5 | 1.6 ± 0.2 a | 1.4 | 69 c | 15 | 5 |
| a-core (Na+) | 1.0 ± 0.04 a | 2.2 | 0.73 ± 0.09 a | 1.0 | 57 c | - | 1 |
| a-coreTT (K+) | 6.1 ± 0.4 a | 1.8 | 1.9 ± 0.3 a | 1.3 | 63.5 c | 18 | - |
| TBA (K+) | 3.5 ± 0.2 a | 1.4 | 0.49 ± 0.04 a | 1.1 | 50.5 c | 11 | - |
| c-kit (K+) | 1.9 ± 0.18 a | 1.4 | 0.74 ± 0.05 a | 1.7 | 54 d | 11 | - |
| c-myc (K+) | 4.0 ± 0.3 a | 1.7 | 1.5 ± 0.2 a | 1.6 | 70 e | 15 | - |
| dsDNA (K+) | 0.037 b | - | 0.60 ± 0.04 a | 2.8 | 49 c | 0.3 | 12 |
| dsDNA (Na+) | 0.037 b | - | 0.60 ± 0.08 a | 3.0 | 49 c | 0.3 | 12 |
2.2. Circular Dichroism (CD) Studies

2.3. Thermal Melting Studies

2.4. TRAP Assay
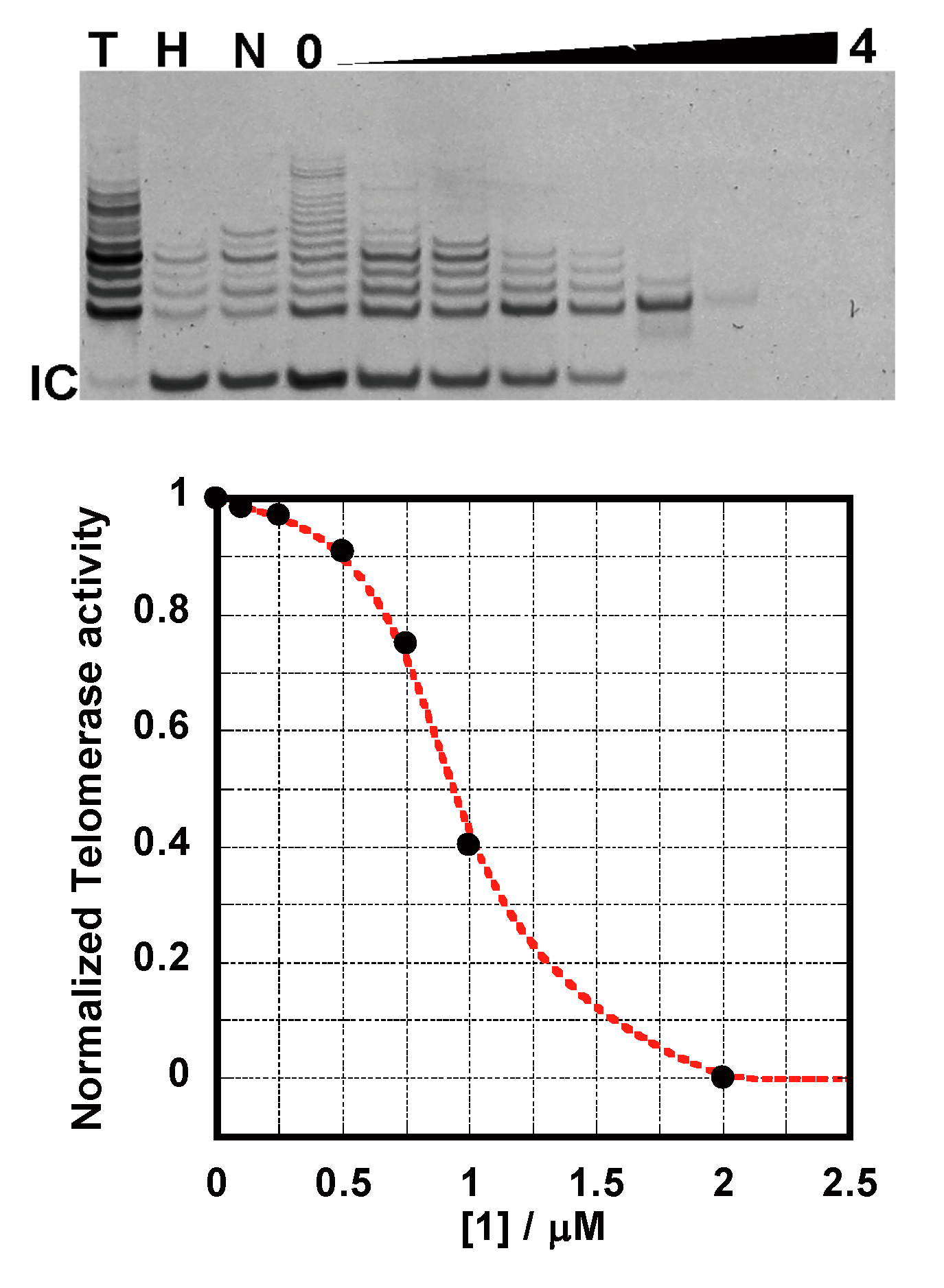
2.5. FRET-Melting Assay

2.6. Computer Modeling
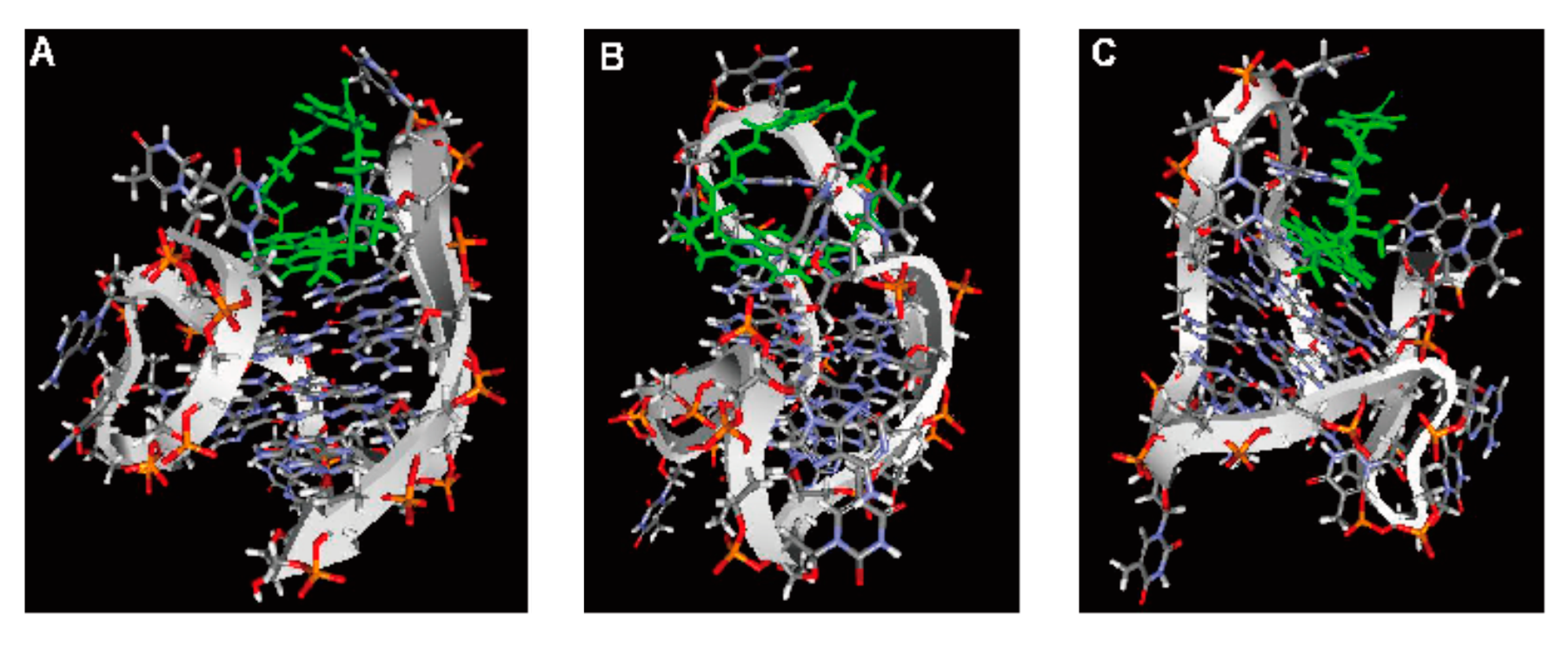
3. Experimental Section
3.1. Materials
3.2. UV-Vis Titration Experiments
3.3. Circular Dichroism (CD) Spectral Measurements
3.4. Thermal Melting Experiments
3.5. TRAP Assay Experiments
3.6. FRET-Melting Assay
3.7. Computer Modeling
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Neidle, S. Therapeutic Applications of Quadruplex Nucleic Acid, 1st ed.; Academic Press: London, UK, 2012; pp. 1–15. [Google Scholar]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Lu, Y.; Tan, J.; Huang, Z.; Wong, K.; Gu, L. G-Quadruplexes: Targets in Anticancer Drug Design. Chem. Med. Chem. 2008, 3, 690–713. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L. Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.M.; Neidle, S.; Parkinson, G.N. A structural analysis of G-quadruplex/ligand interactions. Biochimie 2011, 93, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Monchaud, D.; Teulade-Fichou, M.P. A hitchhiker’s guide to G-quadruplex ligands. Org. Biomol. Chem. 2008, 6, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.T. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew. Chem. Int. Ed. Engl. 2004, 43, 668–698. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.C.; Ulven, T. Macrocyclic G-Quadruplex Ligands. Curr. Med. Chem. 2010, 17, 3438–3448. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, I.; Sato, S.; Juskowiak, B.; Takenaka, S. Interactions of cyclic and non-cyclic naphthalene diimide derivatives with different nucleic acids. Bioorg. Med. Chem. 2014, 22, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Hampel, S.M.; Sidibe, A.; Gunaratnam, M.; Riou, J.; Neidle, S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorganic Med. Chem. Lett. 2010, 20, 6459–6463. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; Minarini, A.; Tumiatti, V.; Moraca, F.; Parrotta, L.; Alcaro, S.; Rigo, R.; Sissi, C.; Gunaratnam, M.; Ohnmacht, S.A.; et al. Macrocyclic naphthalene diimides as G-quadruplex binders. Bioorg. Med. Chem. 2015, in press. [Google Scholar]
- Prato, G.; Silvent, S.; Saka, S.; Lamberto, M.; Kosenkov, D. Thermodynamics of Binding of Di- and Tetrasubstituted Naphthalene Diimide Ligands to DNA G-Quadruplex. J. Phys. Chem. B 2015, 119, 3335–3347. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural Basis for Telomeric G-Quadruplex Targeting by Naphthalene Diimide Ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef] [PubMed]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-Based Design and Evaluation of Naphthalene Diimide G-Quadruplex Ligands As Telomere Targeting Agents in Pancreatic Cancer Cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef] [PubMed]
- Nadai, M.; Doria, F.; Antonio, M.D.; Sattin, G.; Germani, L.; Percivalle, C.; Palumbo, M.; Richter, S.N.; Freccero, M. Naphthalene diimide scaffolds with dual reversible and covalent interaction properties towards G-quadruplex. Biochimie 2011, 93, 1328–1340. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Oppi, A.; Manoli, F.; Botti, S.; Kandoth, N.; Grande, V.; Manet, I.; Freccero, M. A naphthalene diimide dyad for fluorescence switch-on detection of G-quadruplexes. Chem. Commun. 2015, 51, 9105–9108. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Nadai, M.; Folini, M.; Scalabrin, M.; Germani, L.; Sattin, G.; Mella, M.; Palumbo, M.; Zaffaroni, N.; Fabris, D.; et al. Targeting Loop Adenines in G-Quadruplex by a Selective Oxirane. Chem. Eur. J. 2013, 19, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Doria, F.; Nadai, M.; Folini, M.; Antonio, M.D.; Germani, L.; Percivalle, C.; Sissi, C.; Zaffaroni, N.; Alcaro, S.; Artese, A.; et al. Hybrid ligand-alkylating agents targeting telomeric G-quadruplex Structures. Org. Biomol. Chem. 2012, 10, 2798–2806. [Google Scholar] [CrossRef] [PubMed]
- Antonio, M.D.; Folini, M.; Richter, S.N.; Bertipaglia, C.; Mella, M.; Sissi, C.; Manlio Palumbo, M.; Freccero, M. Quinone Methides Tethered to Naphthalene Diimides as Selective G-Quadruplex Alkylating Agents. J. Am. Chem. Soc. 2009, 131, 13132–13141. [Google Scholar] [CrossRef] [PubMed]
- Nadai, M.; Doria, F.; Germani, L.; Richter, S.N.; Freccero, M. A Photoreactive G-Quadruplex Ligand Triggered by Green Light. Chem. Eur. J. 2015, 21, 2330–2334. [Google Scholar] [CrossRef] [PubMed]
- Esaki, Y.; Islam, M.M.; Fujii, S.; Sato, S.; Takenaka, S. Design of tetraplex specific ligands: Cyclic naphthalene diimide. Chem. Commun. 2014, 50, 5967–5969. [Google Scholar] [CrossRef] [PubMed]
- Cian, A.D.; Guittat, L.; Kaiser, M.; Sacca, B.; Amrane, S.; Bourdoncle, A.; Teulade-Fichou, M.P.; Alberti, P.; Lacroix, L.; Mergny, J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [PubMed]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [PubMed]
- Hansel, R.; Lohr, F.; Trantirkova, S.F.; Bamberg, E.; Trantirek, L.; Dotsch, V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011, 39, 5768–5775. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.X.; Carver, M.; Hurley, L.H.; Yang, D.Z. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133, 17673–17680. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an Unprecedented G-Quadruplex Scaffold in the Human c-kit Promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Maiti, S. Effect of Loop Orientation on Quadruplex-TMPyP4 Interaction. J. Phys. Chem. B 2008, 112, 8151–8159. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roet, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.; Gargallo, R.; Eritja, R.; Jaumot, J. Study of the interaction between the G-quadruplex-forming thrombin-binding aptamer and the porphyrin 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)-21,23H-porphyrin tetratosylate. Anal. Biochem. 2008, 379, 8–15. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; vonHippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Benesi, H.A.; Hildebrand, J.H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 1949, 71, 2703–2707. [Google Scholar] [CrossRef]
- Islam, M.M.; Fujii, S.; Sato, S.; Okauchi, T.; Takenaka, S. Thermodynamics and Kinetic Studies in the Binding Interaction of Cyclic Naphthalene Diimide Derivatives with Double Stranded DNAs. Bioorganic Med. Chem. 2015, in press. [Google Scholar]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the Human Telomere in K+ Solution: An Intramolecular (3 + 1) G-Quadruplex Scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhang, D.; Chen, W.; Zhang, M.; Zhou, Y.; Zhou, X. Cationic N-confused porphyrin derivative as a better molecule scaffold for G-quadruplex recognition. Bioorganic Med. Chem. 2010, 18, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Nagatoishi, S.; Tanaka, Y.; Tsumoto, K. Circular dichroism spectra demonstrate formation of the thrombin-binding DNA aptamer G-quadruplex under stabilizing-cation-deficient conditions. Biochem. Biophys. Res. Commun. 2007, 352, 812–817. [Google Scholar] [CrossRef] [PubMed]
- Dash, J.; Shirude, P.S.; Hsu, S.D.; Balasubramanian, S. Diarylethynyl Amides That Recognize the Parallel Conformation of Genomic Promoter DNA G-Quadruplexes. J. Am. Chem. Soc. 2008, 130, 15950–15956. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, Y.; Zhang, M. Synthesis and binding studies of novel di-substituted phenanthroline compounds with genomic promoter and human telomeric DNA G-quadruplexes. Org. Biomol. Chem. 2013, 11, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, V.; Harikrishna, S.; Jagadeeswaran, C.; Halder, K.; Pradeepkumar, P.I. Selective G-quadruplex DNA Stabilizing Agents Based on Bisquinolinium and Bispyridinium Derivatives of 1,8-Naphthyridine. J. Org. Chem. 2012, 77, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Hayden, K.L.; Graves, D.E. Addition of Bases to the 5′-end of Human Telomeric DNA: Influences on Thermal Stability and Energetics of Unfolding. Molecules 2014, 19, 2286–2298. [Google Scholar] [CrossRef] [PubMed]
- Ragazzon, P.; Chaires, J.B. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods 2007, 43, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Fichou, M.P.T.; Carrasco, C.; Guittat, L.; Bailly, C.; Alberti, P.; Mergny, J.L.; David, A.; Lehn, J.M.; Wilson, W.D. Selective Recognition of G-Quadruplex Telomeric DNA by a Bis(quinacridine) Macrocycle. J. Am. Chem. Soc. 2003, 125, 4732–4740. [Google Scholar] [CrossRef] [PubMed]
- Tanious, F.A.; Yen, S.F.; Wilson, W.D. Kinetic and Equilibrium Analysis of a Threading Intercalation Mode: DNA Sequence and Ion Effects. Biochemistry 1991, 30, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 1 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.M.; Fujii, S.; Sato, S.; Okauchi, T.; Takenaka, S. A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative. Molecules 2015, 20, 10963-10979. https://doi.org/10.3390/molecules200610963
Islam MM, Fujii S, Sato S, Okauchi T, Takenaka S. A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative. Molecules. 2015; 20(6):10963-10979. https://doi.org/10.3390/molecules200610963
Chicago/Turabian StyleIslam, Md. Monirul, Satoshi Fujii, Shinobu Sato, Tatsuo Okauchi, and Shigeori Takenaka. 2015. "A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative" Molecules 20, no. 6: 10963-10979. https://doi.org/10.3390/molecules200610963
APA StyleIslam, M. M., Fujii, S., Sato, S., Okauchi, T., & Takenaka, S. (2015). A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative. Molecules, 20(6), 10963-10979. https://doi.org/10.3390/molecules200610963






