Protein Tyrosine Phosphatase 1B Inhibitors from the Roots of Cudrania tricuspidata
Abstract
:1. Introduction
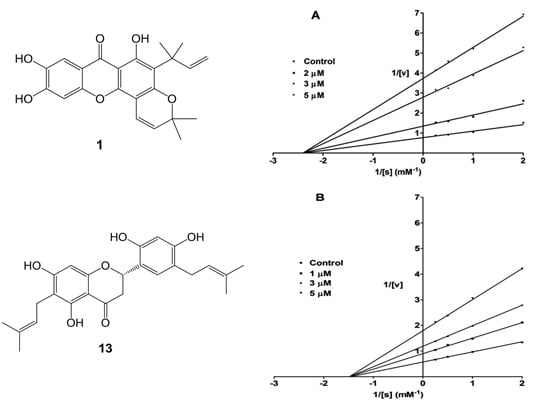
2. Results and Discussion

| Compounds | PTP1B Inhibitory Effects a (IC50 Values = μM) |
|---|---|
| 1 | 2.0 ± 0.4 |
| 2 | 3.0 ± 0.6 |
| 3 | 3.0 ± 0.3 |
| 4 | 4.3 ± 0.6 |
| 5 | 4.6 ± 0.8 |
| 6 | 3.8 ± 0.5 |
| 7 | 1.9 ± 0.4 |
| 8 | 2.8 ± 0.6 |
| 9 | 3.5 ± 0.7 |
| 10 | n.d. |
| 11 | n.d. |
| 12 | n.d. |
| 13 | 5.7 ± 1.5 |
| 14 | 12.3 ± 2.2 |
| 15 | 9.4 ± 2.9 |
| 16 | 13.6 ± 3.3 |
| Ursolic acid b | 3.8 ± 0.4 |

3. Experimental Section
3.1. General
3.2. Plant Material
3.3. Extraction and Isolation
3.4. PTP1B Inhibitory Activity Assay
4. Conclusions
Supplementary Data
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fischer, E.H.; Charbonneau, H.; Tonks, N.K. Protein tyrosine phosphatases: A diverse family of intracellular and transmembrane enzymes. Science 1991, 253, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y. Protein tyrosine phosphatases: Prospects for therapeutics. Curr. Opin. Chem. Biol. 2001, 5, 416–423. [Google Scholar] [CrossRef]
- Andersen, J.N.; Jansen, P.G.; Echwald, S.M.; Mortensen, O.H.; Fukada, T.; Del Vecchio, R.; Tonks, N.K.; Moller, N.P. A genomic perspective on protein tyrosine phosphatases: Gene structure, pseudogenes, and genetic disease linkage. FASEB J. 2004, 18, 8–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Lee, S.Y. PTP1B inhibitors as potential therapeutics in the treatment of type 2 diabetes and obesity. Expert Opin. Investig. Drugs 2003, 12, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. PTP1B: From the sidelines to the front lines! FEBS Lett. 2003, 546, 140–148. [Google Scholar] [CrossRef]
- Asante-Appiah, E.; Kennedy, B.P. Protein tyrosine phosphatases: The quest for negative regulators of insulin action. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E663–E670. [Google Scholar] [CrossRef] [PubMed]
- Zabolotny, J.M.; Bence-Hanulec, K.K.; Stricker-Krongrad, A.; Haj, F.; Wang, Y.; Minokoshi, Y.; Kim, Y.B.; Elmquist, J.K.; Tartaglia, L.A.; Kahn, B.B.; et al. PTP1B regulates leptin signal transduction in vivo. Dev. Cell 2002, 2, 489–495. [Google Scholar] [CrossRef]
- Bence, K.K.; Delibegovic, M.; Xue, B.; Gorgun, C.Z.; Hotamisligil, G.S.; Neel, B.G.; Kahn, B.B. Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat. Med. 2006, 12, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Fantus, I.G. Inhibition of the protein tyrosine phosphatase PTP1B: Potential therapy for obesity, insulin resistance and type-2 diabetes mellitus. Best Pract. Res. Clin. Endoc. Metab. 2007, 21, 621–640. [Google Scholar] [CrossRef] [PubMed]
- Elchebly, M.; Payette, P.; Michaliszyn, E.; Cromlish, W.; Collins, S.; Loy, A.L.; Normandin, D.; Cheng, A.; Himms-Hagen, J.; Chan, C.C.; et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999, 283, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Klaman, L.D.; Boss, O.; Peroni, O.D.; Kim, J.K.; Martino, J.L.; Zabolotny, J.M.; Moghal, N.; Lubkin, M.; Kim, Y.B.; Sharpe, A.H.; et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 2000, 20, 5479–5489. [Google Scholar] [CrossRef] [PubMed]
- Zinker, B.A.; Rondinone, C.M.; Trevillyan, J.M.; Gum, R.J.; Clampit, J.E.; Waring, J.F.; Xie, N.; Wilcox, D.; Jacobson, P.; Frost, L.; et al. PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose, and improves insulin sensitivity in diabetic mice. Proc. Natl. Acad. Sci. USA 2002, 99, 11357–11362. [Google Scholar] [CrossRef] [PubMed]
- Echwald, S.M.; Bach, H.; Vestergaard, H.; Richelsen, B.; Kristensen, K.; Drivsholm, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. A P387L variant in protein tyrosine phosphatase-1B (PTP-1B) is associated with type 2 diabetes and impaired serine phosphorylation of PTP-1B in vitro. Diabetes 2002, 51, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Di Paola, R.; Frittitta, L.; Miscio, G.; Bozzali, M.; Baratta, R.; Centra, M.; Spampinato, D.; Santagati, M.G.; Ercolino, T.; Cisternino, C.; et al. A variation in 3′ UTR of hPTP1B increases specific gene expression and associates with insulin resistance. Am. J. Hum. Genet. 2002, 70, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Mok, A.; Cao, H.; Zinman, B.; Hanley, A.J.; Harris, S.B.; Kennedy, B.P.; Hegele, R.A. A single nucleotide polymorphism in protein tyrosine phosphatase PTP-1B is associated with protection from diabetes or impaired glucose tolerance in Oji-Cree. J. Clin. Endocrinol. Metab. 2002, 87, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Bjorge, J.D.; Fujita, D.J. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer Res. 2007, 67, 10129–10137. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.G.; Dube, N.; Read, M.; Penney, J.; Paquet, M.; Han, Y.; Kennedy, B.P.; Muller, W.J.; Tremblay, M.L. Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nat. Genet. 2007, 39, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K.; Muthuswamy, S.K. A brake becomes an accelerator: PTP1B—A new therapeutic target for breast cancer. Cancer Cell 2007, 11, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.H. The Medicinal Plants of Korea; Kyo-Hak Publishing Co.: Seoul, Korea, 2000; pp. 68–69. [Google Scholar]
- Lee, B.W.; Lee, J.H.; Lee, S.T.; Lee, H.S.; Lee, W.S.; Jeong, T.S.; Park, K.H. Antioxidant and cytotoxic activities of xanthones from Cudrania tricuspidata. Bioorg. Med. Chem. Lett. 2005, 15, 5548–5552. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.W.; Gal, S.W.; Park, K.-M.; Park, K.H. Cytotoxic Xanthones from Cudrania tricuspidata. J. Nat. Prod. 2005, 68, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Hong, S.S.; Han, X.H.; Hwang, J.S.; Lee, D.; Lee, H.; Yun, Y.P.; Kim, Y.; Ro, J.S.; Hwang, B.Y. Prenylated xanthones from the root bark of Cudrania tricuspidata. J. Nat. Prod. 2007, 70, 1207–1209. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.K.; Kim, C.J.; Song, K.S.; Kim, H.M.; Koshino, H.; Uramoto, M.; Yoo, I.D. Cytotoxic benzyl dihydroflavonols from Cudrania tricuspidata. Phytochemistry 1996, 41, 213–216. [Google Scholar] [PubMed]
- Lee, I.K.; Kim, C.J.; Song, K.S.; Kim, H.M.; Yoo, I.D.; Koshino, H.; Esumi, Y.; Uramoto, M. Two benzylated dihydroflavonols from Cudrania tricuspidata. J. Nat. Prod. 1995, 58, 1614–1617. [Google Scholar] [CrossRef]
- Park, K.H.; Park, Y.D.; Han, J.M.; Im, K.R.; Lee, B.W.; Jeong, I.Y.; Jeong, T.S.; Lee, W.S. Anti-atherosclerotic and anti-inflammatory activities of catecholic xanthones and flavonoids isolated from Cudrania tricuspidata. Bioorg. Med. Chem. Lett. 2006, 16, 5580–5583. [Google Scholar] [CrossRef] [PubMed]
- An, R.B.; Sohn, D.H.; Kim, Y.C. Hepatoprotective compounds of the roots of Cudrania tricuspidata on tacrine-induced cytotoxicity in Hep G2 cells. Biol. Pharm. Bull. 2006, 29, 838–840. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Liu, C.C.; Pi, E.H.; Hou, A.J. New Isoprenylated Xanthones from Cudrania tricuspidata. Helv. Chim. Acta 2014, 97, 1683–1688. [Google Scholar] [CrossRef]
- Liang, B.; Li, H.R.; Xu, L.Z.; Yang, S.L. Xanthones from the roots of Cudrania fruticosa Wight. J. Asian Nat. Prod. Res. 2007, 9, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.S.; Hou, A.J.; Zhu, G.F.; Chen, Y.F.; Sun, H.D.; Zhao, Q.S. Cytotoxic isoprenylated xanthones from Cudrania tricuspidata. Bioorg. Med. Chem. 2004, 12, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Curtis-Long, M.J.; Lee, J.W.; Kim, J.H.; Kim, J.Y.; Kang, K.Y.; Lee, W.S.; Park, K.H. Characteristic of neuraminidase inhibitory xanthones from Cudrania tricuspidata. Bioorg. Med. Chem. 2009, 17, 2744–2750. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.H.; Shin, B.; Liu, Q.; Lee, K.Y.; Oh, D.C.; Hwang, B.Y.; Lee, M.K. Antiproliferative prenylated xanthones and benzophenones from the roots of Cudrania tricuspidata in HSC-T6 Cells. J. Nat. Prod. 2014, 77, 2361–2366. [Google Scholar] [CrossRef] [PubMed]
- Hano, Y.; Matsumoto, Y.; Shinohara, K.; Sun, J.Y.; Nomura, T. Structures of Four New Isoprenylated Xanthones, Cudraxanthones L, M, N, and O from Cudrania tricuspidata 1,2. Planta Med. 1991, 57, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, X.; Zhang, P.; Li, Z.L.; Wang, Y. Antiinflammatory constituents from the roots of Smilax bockii warb. Arch. Pharm. Res. 2005, 28, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Ryu, Y.B.; Curtis-Long, M.J.; Ryu, H.W.; Baek, Y.S.; Kang, J.E.; Lee, W.S.; Park, K.H. Tyrosinase inhibitory polyphenols from roots of Morus lhou. J. Agric. Food Chem. 2009, 57, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Yonekawa, M.; Hou, A.J.; Nomura, T.; Sun, H.D.; Uno, J. Antifungal agents from the roots of Cudrania cochinchinensis against Candida, Cryptococcus, and Aspergillus species. J. Nat. Prod. 2003, 66, 1118–1120. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.S.; Hou, A.J.; Zhu, G.F. Isoprenylated xanthones and flavonoids from Cudrania tricuspidata. Chem. Biodivers. 2005, 2, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Shirataki, Y.; Manaka, A.; Yokoe, I.; Komatsu, M. Two prenylflavanones from Euchresta japonica. Phytochemistry 1980, 21, 2959–2963. [Google Scholar] [CrossRef]
- Hano, Y.; Matsumoto, Y.; Shinohara, K.; Sun, J.Y.; Nomura, T. Cudraflavones C and D, two new prenylflavones from the root bark of Cudrania tricuspidata (Carr.) Bur. Heterocycles 1990, 31, 1339–1344. [Google Scholar]
- Nomura, T.; Fukai, T.; Katayanagi, M. Kuwanon A, B, C and oxydihydromorusin, four new flavones from the root bark of the cultivated mulberry tree (Morus alba L.). Chem. Pharm. Bull. 1977, 25, 529–532. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.Y. PTP1B as a drug target: Recent developments in PTP1B inhibitor discovery. Drug Discov. Today 2007, 12, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Bialy, L.; Waldmann, H. Inhibitors of protein tyrosine phosphatases: Next-generation drugs? Angew. Chem. Int. Ed. Engl. 2005, 44, 3814–3839. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.S.; Liang, L.F.; Guo, Y.W. Natural products possessing protein tyrosine phosphatase 1B (PTP1B) inhibitory activity found in the last decades. Acta Pharmacol. Sin. 2012, 33, 1217–1245. [Google Scholar] [CrossRef] [PubMed]
- Son, H.U.; Lee, S.H. Comparison of α-glucosidase inhibition by Cudrania tricuspidata according to harvesting time. Biomed. Rep. 2013, 1, 624–628. [Google Scholar] [PubMed]
- Seo, E.J.; Curtis-Long, M.J.; Lee, B.W.; Kim, H.Y.; Ryu, Y.B.; Jeong, T.S.; Lee, W.S.; Park, K.H. Xanthones from Cudrania tricuspidata displaying potent α-glucosidase inhibition. Bioorg. Med. Chem. Lett. 2007, 17, 6421–6424. [Google Scholar] [CrossRef] [PubMed]
- Park, W.Y.; Ro, J.S.; Lee, K.S. Hypoglycemic effect of Cudrania tricuspidata root bark. Korean J. Pharmacogn. 2001, 32, 248–252. [Google Scholar]
- Quang, T.H.; Ngan, N.T.; Ko, W.; Kim, D.C.; Yoon, C.S.; Sohn, J.H.; Yim, J.H.; Kim, Y.C.; Oh, H. Tanzawaic acid derivatives from a marine isolate of Penicillium sp. (SF-6013) with anti-inflammatory and PTP1B inhibitory activities. Bioorg. Med. Chem. Lett. 2014, 24, 5787–5791. [Google Scholar] [CrossRef] [PubMed]
- Wiesmann, C.; Barr, K.J.; Kung, J.; Zhu, J.; Erlanson, D.A.; Shen, W.; Fahr, B.J.; Zhong, M.; Taylor, L.; Randal, M.; et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat. Struct. Mol. Biol. 2004, 11, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zeng, L.F.; Wu, L.; Yu, X.; Xue, T.; Gunawan, A.M.; Long, Y.Q.; Zhang, Z.Y. Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors. J. Am. Chem. Soc. 2008, 130, 17075–17084. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds 1–16 are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quang, T.H.; Ngan, N.T.T.; Yoon, C.-S.; Cho, K.-H.; Kang, D.G.; Lee, H.S.; Kim, Y.-C.; Oh, H. Protein Tyrosine Phosphatase 1B Inhibitors from the Roots of Cudrania tricuspidata. Molecules 2015, 20, 11173-11183. https://doi.org/10.3390/molecules200611173
Quang TH, Ngan NTT, Yoon C-S, Cho K-H, Kang DG, Lee HS, Kim Y-C, Oh H. Protein Tyrosine Phosphatase 1B Inhibitors from the Roots of Cudrania tricuspidata. Molecules. 2015; 20(6):11173-11183. https://doi.org/10.3390/molecules200611173
Chicago/Turabian StyleQuang, Tran Hong, Nguyen Thi Thanh Ngan, Chi-Su Yoon, Kwang-Ho Cho, Dae Gill Kang, Ho Sub Lee, Youn-Chul Kim, and Hyuncheol Oh. 2015. "Protein Tyrosine Phosphatase 1B Inhibitors from the Roots of Cudrania tricuspidata" Molecules 20, no. 6: 11173-11183. https://doi.org/10.3390/molecules200611173