2.1.1. Establishing Optimal QSAR Models
“Breaking point” method was used to determine the optimal QSAR models of cinnamaldehyde compounds against
Trametes versicolor and
Gloeophyllun trabeum as shown in
Figure 1. The
x-coordinate represents number of descriptors, and the
y-coordinate represents the correlation coefficient
R2 of the corresponding model. As the trend line shows: the correlation coefficient
R2 increased as the number of descriptors increased. When the number of descriptors (
n) was less than 4, the correlation coefficient
R2 increased sharply. The fitting line with high correlation coefficient is 0.997 and 0.9785. When the number of descriptors exceeded 4, the correlation coefficient
R2 increased slightly. The fitting line also had a high correlation coefficient 0.939 and 0.947. According to this method, the breaking point appeared when the number of descriptors was 4 or higher, as shown in
Figure 1. The number of descriptors of the best models should also meet the requirements of multi-linear regression, as evidenced by the number of descriptors (
k) of the optimal models and the sample number (
n) ≥ 3(
k + 1) [
13]. Therefore, the number of descriptors of the optimal QSAR models against
Trametes versicolor and Gloeophyllun trabeum is 4. The value of optimal descriptors is shown in
Table 1 and
Table 2.
The optimal models are shown in
Table 3 and
Table 4, these models had the following statistical characteristics:
R2 = 0.910,
F = 35.32, and
s2 = 0.0093 for
Trametes versicolor;
R2 = 0.926,
F = 43.95, and
s2 = 0.0049 for
Gloeophyllun trabeum.
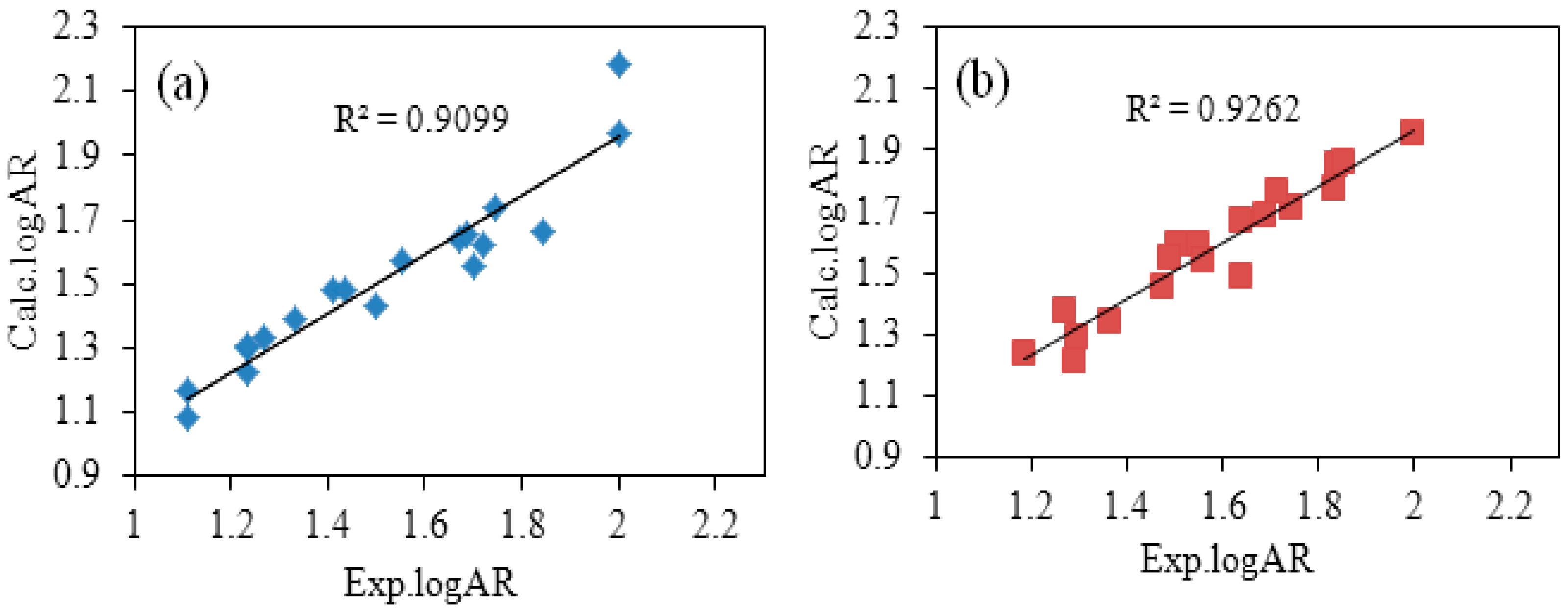
Table 5 shows a comparison between experimental values (Exp.logAR) and calculated values (Calc.logAR). And the plot of Exp.logAR versus Calc.logAR is shown in
Figure 2. The Calc. logAR was calculated according to the optimal QSAR models. There was little difference among Calc.logAR and Exp.logAR, demonstrating that calculated values were close to the experimental values at averages of 0.0661 and 0.0465, respectively, as shown in
Table 5. This miniscule difference indicated that the optimal QSAR models are capable of accurately describing the relationship between chemical structure and bioactivity.
2.1.3. Descriptor Analysis in the Optimal QSAR Models
A
t-test is typically utilized to measure the importance of descriptors in correlation [
15]. According to the
t-test values in
Table 3, the most statistically significant descriptor is the minimum net atomic charge for an H atom, d1. This is a quantum chemical descriptor that indicates the hydrogen-bond and electrostatic interaction between negative ion and positive ion [
16]. In
Table 3, the positive correlation coefficient for d1 demonstrated that increasing the hydrogen-bonding and electrostatic interaction in cinnamaldehyde derivatives led to an increase in antifungal activity against
Trametes versicolor [
17].
The second descriptor was FNSA-3 fractional PNSA (PNSA-3/TMSA), d2 [
18], which is the ratio of PNSA-3 and TMSA that can be computed as follows [
19]:
where TMSA is total area of the molecule and PNSA-3 is the atomic charge weight of the negatively charged molecular surface area [
20]:
where q
A is the partial charge of the atom and S
A is the respective atomic negatively charged solvent-accessible surface area. Both q
A and S
A were computed in Codessa. FNSA-3 is a significant factor on polar active and hydrogen-bond active charges.
The third descriptor was ESP-RPCS relative charged SA (SAMPOS*RPCG) (Quantum-Chemical PC), d3, which is also a quantum chemical descriptor. This descriptor reflects the total molecular surface area and properties of the function group and indicates interactions among polar molecules [
18].
The fourth descriptor was YZ Shadow/YZ Rectangle, d4, a space property descriptor [
21]. The YZ Shadow was calculated by projecting a molecule on the YZ plane, which is related to molecular conformation and molecular orientation. This shape parameter provided a positive indication of the antifungal activity of the cinnamaldehyde derivatives. As the value of descriptor YZ Shadow increased, the antifungal activity of cinnamaldehyde derivatives against
Trametes versicolor also increased.
As shown in
Table 4, the most statistically significant descriptor was the ESP minimum net atomic charge for an H atom, d1 for QSAR model against
Gloeophyllun trabeum. The second most important descriptor was ESP-RPCS Relative positive charged SA(SAMPOS*RPCG) (Quantum-Chemical PC), d5, which is similar to ESP-RPCS Relative charged SA (SAMPOS*RPCG) (Quantum-Chemical PC), d3. It is the result of the partial positive charged surface multiplied by the relative positive charge [
18]. The third and fourth most important descriptors were FNSA-3 (PNSA-3/TMSA) (Quantum-Chemical PC), d6, and FNSA-3Fractional PNSA (PNSA-3/TMSA), d7. These are quantum chemical descriptors which describe the total molecular surface properties and the functional group as well as the activity of polar molecules [
19].
2.1.4. Designing the New Compound with High Bioactivity, Calculating Its AR
Two cinnamaldehyde amino acid Schiff base compounds with satisfactory predicted activities were selected to synthesize and test their antifungal activity, the structures of those two designed compounds were shown in
Figure 3. The chemical structures of new compounds were confirmed by
1H-NMR, IR, MS, HPLC, purity and melting point. The antifungal activity of new compounds was tested by the same method described in
Section 3.2.1, and antifungal activity ratio (
AR) of two designed compounds were listed in
Table 7.
Cinnamaldehyde glutamic acid potassium Schiff base (Compound A). Orange powder; m.p. 233.7–236.5 °C; purity: 91.2817%; IR (cm−1): 1631 (C=O), 1588 (C=N, Carom=Carom), 1492 (Carom=Carom), 754 (Ar-H), 689 (Ar-H); 1H-NMR (400 MHz, D2O) δ 7.83 (t, J = 10.0 Hz, 1H, CH=N-), 7.35 (dd, J = 12.3, 10.8 Hz, 2H, Ar-H), 7.19–7.10 (m, 3H, Ar-H), 6.89 (d, J = 16.0 Hz, 1H, CH=C-), 6.82–6.71 (m, 1H, C=CH-), 3.53 (dd, J = 8.5, 4.9 Hz, 1H, -CH-), 2.10–2.00 (m, 1H, -CH–C), 2.00–1.90 (m, 2H, -CH2-COOK), 1.89–1.80 (m, 1H, -CH-); MS m/z calcd. for C14H13K2NO4 337.0. [M + H]+, found [M + K]+ 376.2.
P-Methoxy cinnamaldehyde glutamic acid potassium Schiff base (Compound B). Orange powder; m.p. 241.4–244.5 °C; purity: 83.749%; IR (cm−1): 1633 (C=O), 1589 (C=N, Carom=Carom), 1520 (Carom=Carom), 816 (Ar-H); 1H-NMR (400 MHz, D2O) δ 8.02 (d, J = 9.0 Hz, 1H, CH=N-), 7.52–7.46 (m, 2H, Ar-H), 7.04 (d, J = 15.9 Hz, 1H, Ar-H), 6.95–6.90 (m, 2H, CH=C-), 6.83 (dd, J = 15.9, 9.0 Hz, 1H, C=CH-), 3.81 (s, 3H, Ar-OCH3), 3.71 (dd, J = 8.6, 5.0 Hz, 1H, -CH-COOK), 2.31–2.24 (m, 1H, -CH-), 2.19–2.10 (m, 2H, COOK-CH2-), 2.04 (d, J = 13.0 Hz, 1H, -CH-); MS m/z calcd. for C15H15K2NO5 367.0. [M + H]+, found [M + K]+ 406.3.
As shown in
Table 7, the designed compounds exhibited better antifungal qualities than the 19 cinnamaldehyde compounds listed in
Figure 4. The
ARGt of the new compounds against
Gloeophyllun trabeum exceeded the
ARTv against
Trametes versicolor, indicated that the new compounds possessed better antifungal properties than cinnamaldehyde alone. Additionally, the antifungal activity of the new compounds against
Gloeophyllun trabeum significantly exceeded the AR of cinnamaldehyde alone. Concerning the experimental logAR and calculated logAR from optimized models, the experimental value was close to the calculated value for both compounds against both fungi. The smallest error was 0.0155 for Compound A against
Gloeophyllun trabeum. This suggested that the QSAR model against
Gloeophyllun trabeum exhibited stronger predictability and stability, with a higher correlation coefficient (
R2 = 0.926) and better validation results than models against
Trametes versicolor (
R2 = 0.910).