Synthesis and Structural Investigation of New Bio-Relevant Complexes of Lanthanides with 5-Hydroxyflavone: DNA Binding and Protein Interaction Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis
- L = C15H9O3, deprotonated 5-hydroxyflavone
- The metal complexes were assigned the following codes, that will be used further in this article: (1), (2), (3), (4); 5-hydroxyflavone will be used as 5-HOF
2.2. Physicochemical Characterization
2.2.1. IR Spectra
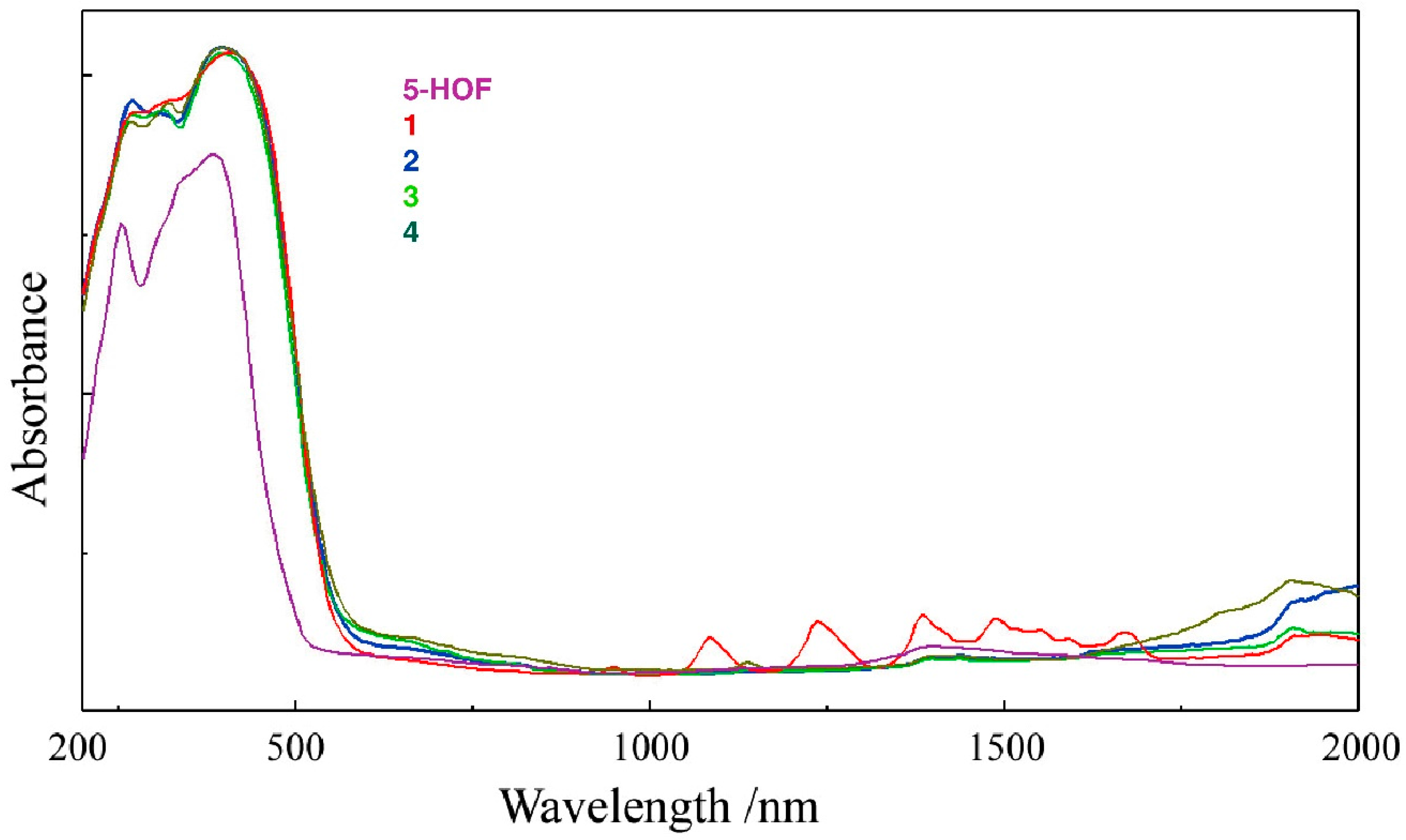
2.2.2. UV-Vis-NIR Spectra
2.2.3. Mass Spectra
2.2.4. Thermal Behavior
2.2.5. Fluorescent Properties
- (i)
- 5-hydroxyflavone does not exhibit fluorescence in the stated conditions;
- (ii)
- at the excitation wavelength of 360 nm, two new bands appeared at 411 and 436 nm in the fluorescence emission spectra of the complexes (1) and (2).
- (iii)
- At the excitation wavelength of 440 nm, a new band appeared at 481 nm in the fluorescence emission spectra of complex 1.
2.2.6. EPR Spectroscopy
2.3. DFT Calculations
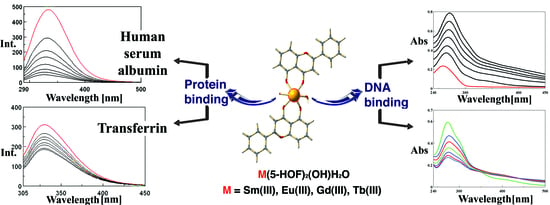
2.4. In Vitro Interactions with Biological Macromolecules
2.4.1. DNA Binding Studies
UV-Vis Spectroscopy Studies
Competitive Binding Studies with Ethidium Bromide Using Fluorescence Spectroscopy
2.4.2. HSA and Tf Binding Studies
Fluorescence Quenching Mechanism
Changes of Tf Conformation Induced by the Interaction with the Tested Compounds
3. Materials and Methods
3.1. Materials
3.2. Analytical Methods
3.3. Synthesis and Characterization of Complexes
3.4. Computational Study
3.5. DNA Binding Studies
3.6. Protein Binding Studies
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Selvaraj, S.; Krishnaswamy, S.; Devashya, V.; Sethuraman, S.; Krishnan, U.M. Flavonoid-metal ion complexes: A novel class of therapeutic agents. Med. Res. Rev. 2014, 34, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Arun, T.R.; Subramanian, R.; Packianathan, S.; Raman, N. Fluorescence Titrations of Bio-relevant Complexes with DNA: Synthesis, Structural Investigation, DNA Binding/Cleavage, Antimicrobial and Molecular Docking Studies. J. Fluoresc. 2015, 25, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Marinic, M.; Piantanida, I.; Rusak, G.; Zinic, M. Interactions of quercetin and its lanthane complex with double stranded DNA/RNA and single stranded RNA: Spectrophotometric sensing of poly G. J. Inorg. Biochem. 2006, 100, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kang, J.; Lu, J.; Li, X.; Tang, J.; Zhang, H.; Zhang, Y. Determination of calf thymus DNA using resonance light-scattering quenching method based on the terbium(III) (Tb3+)/europium(III) (Eu3+)–Quercetin system. J. Lumin. 2009, 129, 906–911. [Google Scholar] [CrossRef]
- Liu, J.Y.; Ren, N.; Zhang, J.J.; Zhang, C.Y.; Song, H.H. Crystal structures, thermal behavior and biological activities of lanthanide compounds with 2,4-dichlorobenzoic acid and 1,10-phenanthroline. Ind. Eng. Chem. Res. 2013, 52, 6156–6163. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, M.; Huang, Y.; Zhang, J.S.; Zhou, G.F.; Zeng, R.Q.; Yang, X.B. Synthesis, characterization, DNA interaction, and antitumor activities of mixed-ligand metal complexes of kaempferol and 1,10-phenanthroline/2,20-bipyridine. Med. Chem. Res. 2014, 23, 2659–2666. [Google Scholar] [CrossRef]
- Kopacz, M.; Woźnicka, E.; Gruszecka, J. Antibacterial activity of morin and its complexes with La(III), Gd(III) and Lu(III) ions. Acta Pol. Pharm. Drug Res. 2005, 62, 65–67. [Google Scholar]
- Xu, W.; Zhou, Y.; Huang, D.; Su, M.; Wang, K.; Hong, M. A highly sensitive and selective fluorescent sensor for detection of Al(3+) using a europium(III) quinolinecarboxylate. Inorg. Chem. 2014, 53, 6497–6499. [Google Scholar] [CrossRef] [PubMed]
- Pusz, J.; Woznicka, E.; Wolowiec, S.; Umbreit, M.H. New solid compounds of Tb(III), Ho(III), Er(III) and Yb(III) with chrysin. J. Therm. Anal. Calorim. 2009, 97, 987–992. [Google Scholar] [CrossRef]
- Ansari, A.A. DFT and 1H-NMR molecular spectroscopic studies on biologically anti-oxidant active paramagnetic lanthanide(III)-chrysin complexes. Main Group Chem. 2008, 7, 43–56. [Google Scholar] [CrossRef]
- Pusz, J.; Wolowiec, S. Solid compounds of Ce(III), Pr(III), Nd(III), and Sm(III) ions with chrysin. J. Therm. Anal. Calorim. 2012, 110, 813–821. [Google Scholar] [CrossRef]
- Ansari, A.A. 1H-NMR, spectroscopic and molecular modeling studies on paramagnetic lanthanide(III)-quercetin complexes. Main Group Chem. 2008, 7, 15–30. [Google Scholar] [CrossRef]
- Ansari, A.A. Paramagnetic NMR shift, spectroscopic and molecular modeling studies of lanthanide(III)-morin complexes. J. Coord. Chem. 2008, 61, 3869–3878. [Google Scholar] [CrossRef]
- Nowak, D.; Woznicka, E.; Kuzniar, A.; Kopacz, M. Magnetism of the lanthanides(III) complexes with some polihydroxyflavones. J. Alloys Compd. 2006, 425, 59–63. [Google Scholar] [CrossRef]
- Woznicka, E.; Kopacz, M.; Umbreit, M.; Klos, J. New complexes of La(III), Ce(III), Pr(III), Nd(III), Sm(III), Eu(III) and Gd(III) ions with morin. J. Inorg. Biochem. 2007, 101, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Rescifina, A.; Zagni, C.; Varrica, M.G.; Pistarà, V.; Corsaro, A. Recent advances in small organic molecules as DNA intercalating agents: Synthesis, activity, and modeling. Eur. J. Med. Chem. 2013, 74, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadi, J.E.N. Molecular aspects on the interaction of quercetin and its metal complexes with DNA. Int. J. Biol. Macromol. 2011, 48, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, S.; Wang, X.; Li, P.; He, Y. A novel quantum dots-based OFF–ON fluorescent biosensor for highly selective and sensitive detection of double-strand DNA. Sens. Actuators B Chem. 2013, 176, 1147–1153. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, J.; Pan, J.; Chen, X.; Wang, J. Spectroscopic studies on the interaction of morin-Eu(III) complex with calf thymus DNA. J. Mol. Struct. 2009, 923, 114–119. [Google Scholar] [CrossRef]
- Li, Z.; Kang, J.; Lu, X. Electrochemical study on behavior of EuMo2 complex and its interaction with DNA. Nucleosides Nucleotides Nucleic Acids 2007, 26, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.D.; Yang, Z.Y.; Wang, Q.; Cai, T.K.; Crewdson, P. Synthesis, characterization, cytotoxic activities, and DNA-binding properties of the La(III) complex with Naringenin Schiff-base. Bioorg. Med. Chem. 2006, 14, 1880–1888. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.T.; Yan, C.Y.; Peng, X.M.; Zhang, S.L.; Rasheed, S.; Geng, R.X.; Zhou, C.H. Synthesis and biological evaluation of α-triazolyl chalcones as a new type of potential antimicrobial agents and their interaction with calf thymus DNA and human serum albumin. Eur. J. Med. Chem. 2014, 71, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Suárez, J.; Shand, T.; Magliozzo, R.S.; Sánchez-Delgado, R.A. Interactions of arene-Ru(II)-chloroquine complexes of known antimalarial and antitumor activity with human serum albumin (HSA) and transferrin. J. Inorg. Biochem. 2011, 105, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Han, R.M.; Sun, X.R.; Li, G.Y.; Yang, Q.F.; Li, Q.; Gai, W.; Zhang, M.; Chen, L.; Yang, G.; et al. The effect of the skeleton structure of flavanone and flavonoid on interaction with transferrin. Bioorg. Med. Chem. Lett. 2013, 23, 6677–6681. [Google Scholar] [CrossRef] [PubMed]
- Valant-Vetschera, K.M.; Bhutia, T.D.; Wollenweber, E. Exudate flavonoids of Primula spp: Structural and biogenetic chemodiversity. Nat. Prod. Commun. 2009, 4, 365–370. [Google Scholar] [PubMed]
- Valant-Vetschera, K.M.; Bhutia, T.D.; Wollenweber, E. Chemodiversity of exudate flavonoids in Dionysia (Primulaceae): A comparative study. Phytochemistry 2010, 71, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Calderone, V.; Chericoni, S.; Martinelli, C.; Testai, L.; Nardi, A.; Morelli, I.; Breschi, M.C.; Martinotti, E. Vasorelaxing effects of flavonoids: Investigation on the possible involvement of potassium channels. Naunyn Schmiedebergs Arch. Pharmacol. 2004, 370, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Nishizaki, Y.; Ishimoto, Y.; Hotta, Y.; Hosoda, A.; Yoshikawa, H.; Akamatsu, M.; Tamura, H. Effect of flavonoids on androgen and glucocorticoid receptors based on in vitro reporter gene assay. Bioorg. Med. Chem. Lett. 2009, 19, 4706–4710. [Google Scholar] [CrossRef] [PubMed]
- Dangleterre, L.; Cornard, J.-P.; Lapouge, C. Spectroscopic and theoretical investigation of the solvent effects on Al(III)–hydroxyflavone complexes. Polyhedron 2008, 27, 1581–1590. [Google Scholar] [CrossRef]
- Sarowar, C.H.; Moran, G.; Willett, G.D. A study of divalent metal cations Cu2+, Zn2+ and Pb2+ attachment to 3-hydroxyflavone, 5-hydroxyflavone and 5-methoxyflavone by nanoelectrospray ionization LTQ Orbitrap mass spectrometry. Int. J. Mass Spectrom. 2013, 333, 44–54. [Google Scholar] [CrossRef]
- Cornard, J.P.; Merlin, J.C. Structural and spectroscopic investigation of 5-hydroxyflavone and its complex with aluminium. J. Mol. Struct. 2001, 569, 129–138. [Google Scholar] [CrossRef]
- Jabeen, E.; Ahmed, S.; Murtaza, I.; Ali, T.; Hameed, S. Radical scavenging propensity of Cu2+, Fe3+ complexes of flavonoids and in vivo radical scavenging by Fe3+-primuletin. Spectrochim. Acta A 2017, 171, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Sathish, R.S.; Goutam, A.R.; Rao, G.N.; Janardhana, C. A fluorescent fluoride ion probe based on a self-organized ensemble of 5-hydroxyflavone-Al(III) complex. Spectrochim. Acta A 2008, 69, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Yuji, H.; Sano, T.; Fujii, H.; Nishio, Y.; Takanashi, H.; Shibata, K.H. Organic light emitting diodes using 3- or 5-hydroxyflavone–metal complexes. Appl. Phys. Lett. 1997, 71, 3338–3340. [Google Scholar]
- Uivarosi, V.; Barbuceanu, S.F.; Aldea, V.; Arama, C.C.; Badea, M.; Olar, R.; Marinescu, D. Synthesis, Spectral and Thermal Studies of New Rutin Vanadyl Complexes. Molecules 2010, 15, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Uivarosi, V.; Badea, M.; Olar, R.; Draghici, C.; Barbuceanu, S.F. Synthesis and Characterization of Some New Complexes of Magnesium (II) and Zinc (II) with the Natural Flavonoid Primuletin. Molecules 2013, 18, 7631–7645. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, K.; Iyyam Pillai, S.; Subramanian, S.P. Design, synthesis and characterization of zinc-3 hydroxy flavone, a novel zinc metallo complex for the treatment of experimental diabetes in rats. Eur. J. Pharmacol. 2012, 680, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Iyyam Pillai, S.; Subramanian, S.P.; Kandaswamy, M.A. Novel insulin mimetic vanadium flavonol complex: Synthesis, characterization and in vivo evaluation in STZ-induced rats. Eur. J. Med. Chem. 2013, 63, 109–117. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.F.V.; De Giovani, W.F. Synthesis, spectral and electrochemical properties of Al(III) and Zn(II) complexes with flavonoids. Spectrochim. Acta A 2005, 61, 1985–1990. [Google Scholar] [CrossRef] [PubMed]
- Sastri, V.S.; Bünzli, J.C.; Rao, V.R.; Rayudu, G.V.S.; Perumareddi, J.R. Spectroscopy of Lanthanide Complexes. In Modern Aspects of Rare Earths and Their Complexes; Sastri, V.S., Bünzli, J.C., Rao, V.R., Rayudu, G.V.S., Perumareddi, J.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2003; p. 619. [Google Scholar]
- Henmi, K.; Hinatsu, Y.; Masaki, N.M. Crystal Structures and Magnetic Properties of Ordered Perovskites Ba2LnNbO6 (Ln=Lanthanide Elements). J. Solid State Chem. 1999, 148, 353–360. [Google Scholar] [CrossRef]
- Szyczewski, A.; Lis, S.; Krzystek, J.; Staninski, K.; Klonkowski, A.; Kruczynski, Z.; Pietraszkiewicz, M. Gadolinium(III) cryptates investigated by multifrequency EPR. J. Alloys Compd. 2008, 451, 182–185. [Google Scholar] [CrossRef]
- Gudasi, K.B.; Shenoy, R.V.; Vadavi, R.S.; Patil, M.S.; Patil, S.A.; Hanchinal, R.R.; Desai, S.A.; Lohithaswa, H. Lanthanide(III) and Yttrium(III) Complexes of Benzimidazole-2-Acetic Acid: Synthesis, Characterisation and Effect of La(III) Complex on Germination of Wheat. Bioinorg. Chem. Appl. 2006, 2006, 75612. [Google Scholar] [CrossRef] [PubMed]
- Mazur, M.; Poprac, P.; Valko, M.; Rhodes, C. ‘U-spectrum’ type of Gd(III) EPR spectra recorded at various stages of TEOS-based sol-gel process. J. Sol-Gel Sci. Technol. 2016, 79, 220–227. [Google Scholar] [CrossRef]
- Essawy, A.A.; Afifi, M.A.; Moustafa, H.; El-Medani, S.M. DFT calculations, spectroscopic, thermal analysis and biological activity of Sm(III) and Tb(III) complexes with 2-aminobenzoic and 2-amino-5-chloro-benzoic acids. Spectrochim. Acta A 2014, 131, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, B.; Zhu, L. DNA binding and oxidative DNA damage induced by a quercetin copper (II) complex. J. Biol. Inorg. Chem. 2009, 14, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Rusak, G.; Piantanida, I.; Masic, L.; Kapuralin, K.; Durgo, K.; Kopjar, N. Spectrophotometric analysis of flavonoids-DNA interactions and DNA damaging/protecting and cytotoxic potential of flavonoids in human peripheral blood lymphocytes. Chem. Biol. Interact. 2010, 188, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Buchtík, R.; Trávníček, Z.; Vančo, J.; Herchel, R.; Dvořák, Z. Synthesis, characterization, DNA interaction and cleavage, and in vitro cytotoxicity of copper(II) mixed-ligand complexes with 2-phenyl-3-hydroxy-4(1H)-quinolinone. Dalton Trans. 2011, 40, 9404–9412. [Google Scholar] [CrossRef] [PubMed]
- Navarra, M.; Hernandez, C.; Colmenares, I.; Hernandez, P.; Fernandez, M.; Sierraalta, A.; Marchan, E. Synthesis and characterization of [Au(dppz)2]Cl3. DNA interaction studies and biological activity against Leishmania(L) mexicana. J. Inorg. Biochem. 2007, 101, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Sarzehi, S.; Chamani, J. Investigation on the interaction between tamoxifen and human holo-transferrin: Determination of the binding mechanism by fluorescence quenching, resonance light scattering and circular dichroism methods. Int. J. Biol. Macromol. 2010, 47, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Varlan, A.; Hillebrand, M. Bovine and Human Serum Albumin Interactions with 3-Carboxyphenoxathiin Studied by Fluorescence and Circular Dichroism Spectroscopy. Molecules 2010, 15, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yuan, W.; Wang, H.; Zhang, Q.; Liu, M.; Yu, K. Synthesis, crystal structure and interaction with DNA and HSA of (N,N′-dibenzylethane-1,2-diamine) transition metal complexes. J. Inorg. Biochem. 2008, 102, 2026–2034. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Xiang, J.; Zhang, Y.; Tang, Y. A spectroscopic and molecular modeling study of sinomenine binding to transferrin. Bioorg. Med. Chem. Lett. 2007, 17, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.B.; Lin, Z.H.; Liu, H.F.; Le, X.Y. A new ternary copper(II) complex derived from 2-(2′-pyridyl)benzimidazole and glycylglycine: Synthesis, characterization, DNA binding and cleavage, antioxidation and HSA interaction. Spectrochim. Acta A 2014, 122, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Yang, G.; Dong, Y.; Zhao, Y.Q.; Sun, X.R.; Chen, L.; Chen, H.B. Studies on the binding of fulvic acid with transferrin by spectroscopic analysis. Spectrochim. Acta A 2015, 137, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shao, C.; Ji, W.; Xiao, M.; Yi, F.; Zhou, T.; Zi, Y. Studies on the Binding Mechanism of VB1 and VB9 with Trypsin. Am. J. Anal. Chem. 2013, 4, 771–775. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A.02; Gaussian: Wallingford, CT, USA, 2009. [Google Scholar]
- Dolg, M.; Stoll, H.; Savin, A.; Preuss, H. Energy-adjusted pseudopotentials for the rare earth elements. Theor. Chem. Acc. 1989, 75, 173–194. [Google Scholar] [CrossRef]
- Alkauskas, A.; Baratoff, A.; Bruder, C. Gaussian Form of Effective Core Potential and Response Function Basis Set Derived from Troullier–Martins Pseudopotential: Results for Ag and Au. J. Phys. Chem. A 2004, 108, 6863–6868. [Google Scholar] [CrossRef]
- Georgieva, I.; Mihaylov, T.; Trendafilova, N. Lanthanide and transition metal complexes of bioactive coumarins: Molecular modeling and spectroscopic studies. J. Inorg. Biochem. 2014, 135, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, M.; Kia, A.F.A.; Duff, B.; Egan, D.A.; Devereux, M.; McClean, S.; Walsh, M.; Trendafilova, N.; Georgieva, I.; Creaven, B.S. Spectroscopic studies, DFT calculations, and cytotoxic activity of novel silver(I) complexes of hydroxy ortho-substituted-nitro-2 H-chromen-2-one ligands and a phenanthroline adduct. J. Inorg. Biochem. 2015, 153, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Isab, A.A.; Alotaibi, M.A.; Saleem, M.; Monim-ul-Mehboob, M.; Ahmad, S.; Georgieva, I.; Trendafilova, N. Synthesis, spectroscopic characterization, DFT calculations and antimicrobial properties of silver(I) complexes of 2,2′-bipyridine and 1,10-phenanthroline. Polyhedron 2016, 115, 212–218. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds (1), (2), (3), (4) are available from Valentina Uivarosi.








| Complex | Step | Thermal Effect | Temperature Range (°C) | Δmexp (%) | Δmcalc (%) |
|---|---|---|---|---|---|
| (1) | 1 | Exothermic | 320–1000 | 72.5 | 72.8 |
| Residue (Sm2O3) | 27.5 | 27.2 | |||
| (2) | 1 | Endothermic | 170–300 | 5.0 | 5.3 |
| 2 | Exothermic | 320–1000 | 72.2 | 72.3 | |
| Residue (Eu2O3) | 22.8 | 22.4 | |||
| (3) | 1 | Endothermic | 240–315 | 2.6 | 2.7 |
| 2 | Exothermic | 325–1000 | 69.9 | 70.1 | |
| Residue (Gd2O3) | 27.5 | 27.2 | |||
| (4) | 1 | Endothermic | 210–310 | 2.5 | 2.7 |
| 2 | Exothermic | 320–1000 | 69.6 | 69.9 | |
| Residue (Tb2O3) | 27.9 | 27.4 |
| Bond Length (A) | |||
| C4-O7 | 1.351 | ||
| C9=O27 | 1.221 | ||
| Bond Angle (°) | |||
| C4-C5-C9 | 122.998 | ||
| O7-C4-C5 | 120.781 | ||
| O27-C9-C5 | 121.151 | ||
| Charge | |||
| C4 | 0.836 | O7 | −0.434 |
| C9 | 0.448 | O27 | −0.469 |
| Total energy (a.u.) | −802.4868 | ||
| Total dipole moment (D) | 4.3673 | ||
| Parameter | Complex | |||
|---|---|---|---|---|
| (1) | (2) | (3) | (4) | |
| Bond Length (A) | ||||
| M-O7 | 2.155 | 2.149 | 2.156 | 2.150 |
| M-O27 | 2.132 | 2.120 | 2.233 | 2.201 |
| M-O29 | 2.167 | 2.151 | 2.252 | 2.259 |
| M-O30 | 2.149 | 2.146 | 2.177 | 2.124 |
| M-O31 | 2.361 | 2.485 | 2.395 | 2.395 |
| M-O58 | - | 2.410 | 2.379 | 2.378 |
| C9-O27 | 1.276 | 1.304 | 1.306 | 1.307 |
| C4-O7 | 1.281 | 1.312 | 1.310 | 1.315 |
| Bond Angle (°) | ||||
| O27-M-O31 | 54.577 | 48.731 | 51.462 | 51.624 |
| O27-M-O7 | 73.051 | 71.572 | 71.167 | 71.512 |
| O7-M-O29 | 111.091 | 105.489 | 97.734 | 97.707 |
| O30-M-O31 | 46.376 | 49.686 | 53.136 | 50.077 |
| O30-M-O29 | 74.919 | 71.119 | 69.736 | 72.046 |
| O29-M-O58 | - | 57.411 | 48.829 | 49.279 |
| O58-M-O7 | - | 50.274 | 51.109 | 50.623 |
| C4-C5-C9 | 122.998 | 120.776 | 119.586 | 121.189 |
| O7-C4-C5 | 120.781 | 120.263 | 121.871 | 120.425 |
| Dihedral Angle | ||||
| M-O27-C9-C5 | −1.366 | −21.069 | −30.917 | −30.948 |
| M-O7-C4-C5 | −3.011 | 18.903 | 18.397 | 18.769 |
| O31-M-O7-C4 | −1.855 | −41.144 | −47.061 | −47.380 |
| M-O7-C4-C3 | 179.599 | −158.486 | −156.593 | −156.435 |
| M-O29-C33-C37 | 164.413 | 136.736 | 139.683 | 145.150 |
| Charge | ||||
| M | 1.079 | 1.186 | 0.887 | 1.076 |
| O7 | −0.705 | −0.585 | −0.541 | −0.556 |
| O27 | −0.611 | −0.377 | −0.395 | −0.344 |
| O29 | −0.639 | −0.468 | −0.383 | −0.396 |
| O30 | −0.568 | −0.529 | −0.592 | −0.556 |
| O31 | −0.501 | −0.521 | −0.416 | −0.549 |
| O58 | − | −0.635 | −0.626 | −0.628 |
| C4 | 0.406 | 0.228 | 0.221 | 0.213 |
| C9 | 0.561 | 0.206 | 0.163 | 0.121 |
| C33 | 0.554 | 0.255 | 0.141 | 0.162 |
| C38 | 0.406 | 0.221 | 0.281 | 0.233 |
| Total energy (a.u.) | −1706.6004 | −2467.6373 | −2522.8014 | −2581.6789 |
| Total dipole moment (D) | 4.3164 | 7.2773 | 6.7800 | 5.6414 |
| Compound | Kb (L∙mol−1) | R2 |
|---|---|---|
| 5-HOF | 1.47 × 104 | 0.9999 |
| (2) | 2.73 × 104 | 0.994 |
| (3) | 2.93 × 104 | 0.9934 |
| (4) | 3.85 × 104 | 0.9981 |
| Compound | Ksv (L∙mol−1) | R2 |
|---|---|---|
| 5-HOF | 2.89 × 104 | 0.9928 |
| (2) | 1.73 × 105 | 0.9959 |
| (3) | 4.13 × 105 | 0.9941 |
| (4) | 1.39 × 105 | 0.9902 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munteanu, A.-C.; Badea, M.; Olar, R.; Silvestro, L.; Dulea, C.; Negut, C.-D.; Uivarosi, V. Synthesis and Structural Investigation of New Bio-Relevant Complexes of Lanthanides with 5-Hydroxyflavone: DNA Binding and Protein Interaction Studies. Molecules 2016, 21, 1737. https://doi.org/10.3390/molecules21121737
Munteanu A-C, Badea M, Olar R, Silvestro L, Dulea C, Negut C-D, Uivarosi V. Synthesis and Structural Investigation of New Bio-Relevant Complexes of Lanthanides with 5-Hydroxyflavone: DNA Binding and Protein Interaction Studies. Molecules. 2016; 21(12):1737. https://doi.org/10.3390/molecules21121737
Chicago/Turabian StyleMunteanu, Alexandra-Cristina, Mihaela Badea, Rodica Olar, Luigi Silvestro, Constanţa Dulea, Constantin-Daniel Negut, and Valentina Uivarosi. 2016. "Synthesis and Structural Investigation of New Bio-Relevant Complexes of Lanthanides with 5-Hydroxyflavone: DNA Binding and Protein Interaction Studies" Molecules 21, no. 12: 1737. https://doi.org/10.3390/molecules21121737
APA StyleMunteanu, A.-C., Badea, M., Olar, R., Silvestro, L., Dulea, C., Negut, C.-D., & Uivarosi, V. (2016). Synthesis and Structural Investigation of New Bio-Relevant Complexes of Lanthanides with 5-Hydroxyflavone: DNA Binding and Protein Interaction Studies. Molecules, 21(12), 1737. https://doi.org/10.3390/molecules21121737