Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa
Abstract
:1. Introduction
2. Results and Discussion
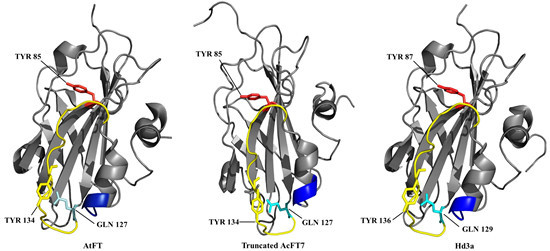
2.1. Search for Onion FT Genes and Phylogenetic Analyses


2.2. Gene Expression Correlates with Bulb Initiation

2.3. Photoperiod Control in Bulb Formation

2.4. FT Gene Expression in Drought Stress

3. Materials and Methods
3.1. Plant Material
3.2. Sequence Analysis
3.3. Analysis of Gene Expression
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tan, F.C.; Swain, S.M. Genetics of flower initiation and development in annual and perennial plants. Physiol. Plant. 2006, 128, 8–17. [Google Scholar] [CrossRef]
- Wilkie, J.D.; Sedgley, M.; Olesen, T. Regulation of floral initiation in horticultural trees. J. Exp. Bot. 2008, 59, 3215–3228. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in arabidopsis. Cell 2010, 141, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L. Environmental physiology of the onion: Towards quantitative models for the effects of photoperiod, temperature and irradiance on bulbing, flowering and growth. Acta Hortic. 1997, 433, 347–373. [Google Scholar] [CrossRef]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Grandy, D.K.; Hanneman, E.; Bunzow, J.; Shih, M.; Machida, C.A.; Bidlack, J.M.; Civelli, O. Purification, cloning, and tissue distribution of a 23-kDa rat protein isolated by morphine affinity chromatography. Mol. Endocrinol. 1990, 4, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Schoentgen, F.; Jolles, P. From structure to function: Possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett. 1995, 369, 22–26. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Goto, K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 2011, 23, 3172–3184. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Bradley, D.J.; Coen, E.S. Separation of shoot and floral identity in Arabidopsis. Development 1999, 126, 1109–1120. [Google Scholar] [PubMed]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER of FT and TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Kobayashi, Y.; Monna, Y.; Sasaki, L.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef] [PubMed]
- Faure, S.; Higgins, J.; Turner, A.; Laurie, D.A. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 2007, 176, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Kawahigashi, H.; Ando, T.; Tonooka, T.; Handa, H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009, 149, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.P.; Minow, M.A.A.; Chalfun-Júnior, A.; Colasanti, J.; Chalfun-Junior, A.; Colasanti, J. Putative sugarcane FT/TFL1 genes delay flowering time and alter reproductive architecture in Arabidopsis. Front. Plant Sci. 2014, 5, 221. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Kawazu, Y.; Fujiyama, R.; Honda, I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 2011, 168, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Yin, J.; Wang, B.; Zhang, Y.; Yang, Q. Molecular Cloning and Expression Analysis of a FT Homologous Gene from Solanum tuberosum. Agric. Sci. China 2010, 9, 1133–1139. [Google Scholar] [CrossRef]
- Li, C.; Luo, L.; Fu, Q.; Niu, L.; Xu, Z.-F. Identification and Characterization of the FT/TFL1 Gene Family in the Biofuel Plant Jatropha curcas. Plant Mol. Biol. Report. 2014, 33, 326–333. [Google Scholar] [CrossRef]
- Lv, L.; Duan, J.; Xie, J.; Wei, C.; Liu, Y.; Liu, S.; Sun, G. Isolation and characterization of a FLOWERING LOCUS T homolog from pineapple (Ananas comosus (L.) Merr). Gene 2012, 505, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Li, X.; Qin, D.; Guo, F.; Wu, C.; Miao, L.; Sun, C. Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol. Biochem. 2012, 58, 98–105. [Google Scholar] [PubMed]
- Pin, P.A.; Benlloch, R.; Bonnet, D.; Wremerth-Weich, E.; Kraft, T.; Gielen, J.J.L.; Nilsson, O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 2010, 330, 1397–1400. [Google Scholar] [CrossRef] [PubMed]
- Winterhagen, P.; Tiyayon, P.; Samach, A.; Hegele, M.; Wunsche, J.N. Isolation and characterization of FLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol. Biochem. 2013, 71, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Baldwin, S.; Kenel, F.; McCallum, J.; Macknight, R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 2013, 4, 2884. [Google Scholar] [CrossRef] [PubMed]
- Lifschitz, E.; Eviatar, T.; Rozman, A.; Shalit, A.; Goldshmidt, A.; Amsellem, Z.; Alvarez, J.P.; Eshed, Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 6398–6403. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L. Physiology of Crop Growth and Bulbing; CRC Press: Boca Raton, FL, USA, 1990; pp. 53–88. [Google Scholar]
- DiAlign: Local Multiple Alignment. Available online: http://www.genomatix.de/cgi-bin/dialign/dialign.pl (accessed on 24 August 2015).
- Garnier, J.; Gibrat, J.F.; Robson, B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzym. 1996, 266, 540–553. [Google Scholar]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; McGonigle, B.; Muszynski, M.G. Beyond flowering time: pleiotropic function of the maize flowering hormone florigen. Plant Signal Behav. 2011, 6, 1267–1270. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Gutfinger, T.; Hareven, D.; Ben-Naim, O.; Ron, N.; Adir, N.; Lifschitz, E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 2001, 13, 2687–2702. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Harig, L.; Beinecke, F.A.; Oltmanns, J.; Muth, J.; Müller, O.; Rüping, B.; Twyman, R.M.; Fischer, R.; Prüfer, D.; Noll, G.A. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 2012, 72, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A Genomic and Expression Compendium of the Expanded PEBP Gene Family from Maize. Plant Physiol. 2007, 146, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Mettananda, R.; Fordham, K.A. The effects of 12 and 16 hours daylength treatments on the onset of bulbing in 21 onion cultivars (Allium cepa L.) and its application to screening germplasm for use in the tropics. J. Hortic. Sci. Biotechnol. 1997, 72, 981. [Google Scholar] [CrossRef]
- Lancaster, J.E.; Triggs, C.M.; de Ruiter, J.M.; Gandar, P.W. Bulbing in onions: Photoperiod and temperature requirements and prediction of bulb size and maturity. Ann. Bot. 1996, 78, 423. [Google Scholar] [CrossRef]
- Mondal, M.F.; Brewster, J.L.; Morris, G.E. L.; Butler, H.A. Bulb Development in Onion (Allium cepa L.) II. The Influence of Red: Far-red Spectral Ratio and of Photon Flux Density. Ann. Bot. 1986, 58, 197–206. [Google Scholar]
- Sobeih, W.Y.; Wright, C.J. The Photoperiodic Regulation Of Bulbing In Onions (Allium-Cepa L).2. Effects of Plant-Age And Size. J. Hortic. Sci. 1986, 61, 337–341. [Google Scholar]
- Taylor, A.; Massiah, A.J.; Thomas, B. Conservation of Arabidopsis thaliana photoperiodic flowering time genes in onion (Allium cepa L.). Plant Cell Physiol. 2010, 51, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- Shalit, A.; Rozman, A.; Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y.; Lifschitz, E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 2009, 106, 8392–8397. [Google Scholar] [CrossRef] [PubMed]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Böhlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Ono, N.; Hayashi, Y.; Morimoto, S.; Nakamura, S.; Soda, M.; Kato, Y.; Ohnishi, M.; Nakano, T.; Inoue, S.; Shimazaki, K. FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 2011, 21, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Lercari, B. Changes in invertase activities during the photoperiodically induced bulb formation of onion (Allium cepa L.). Physiol. Plant. 1982, 54, 480–484. [Google Scholar] [CrossRef]
- Wickramasinghe, U.L.; Wright, C.J.; Currah, L. Bulbing responses of two cultivars of red tropical onions to photoperiod, light integral and temperature under controlled growth conditions. J. Hortic. Sci. Biotechnol. 2000, 75, 304–311. [Google Scholar]
- González, M.I. Effect of sowing date on the production of three storage varieties of onion in the Eighth Region of Chile. Acta Hortic. 1997, 433, 549–554. [Google Scholar] [CrossRef]
- Randle, W.M. Onion Flavor Chemistry and Factors Influencing Flavor Intensity. In Flavor Chemistry and Antioxidant Properties; Risch, S.J., Ho, C.-T., Eds.; ACS Symposium Series Vol. 660: Washington, DC, USA, 1997; pp. 41–52. [Google Scholar]
- Randle, W.M.; Lancaster, J.E. Sulphur compounds in alliums in relation to flavour quality. In Allium Crop Science: Recent Advances; Rabinowitch, H.D., Currah, L., Eds.; CAB International: Wallingford, UK, 2002; pp. 329–356. [Google Scholar]
- Kahane, R.; Rancillac, M.; Serve, B.T. de la Long-term multiplication of onion (Allium cepa L.) by cyclic shoot regeneration in vitro. Plant Cell. Tissue Organ Cult. 1992, 28, 281–288. [Google Scholar] [CrossRef]
- Kahane, R.; Schweisguth, B.; Rancillac, M. Trophic versus environmental factors controlling in vitro bulb formation in onion and garlic micropropagated plants. First Int. Symp. Edible Alliaceae. Acta Hortic. 1997, 433, 435–443. [Google Scholar] [CrossRef]
- Takagi, H. Garlic (Allium sativum L.). In Onions and allied crops. Vol. III. Biochemistry, Food Science and Minor Crops; Rabinowitch, H.D., Brewster, J.L., Eds.; CRC Press: Boca Raton, FL, USA, 1990; Volume 3, pp. 109–116. [Google Scholar]
- Nagakubo, T.; Nagasawa, A.; Ohkawa, H. Micropropagation of garlic through in vitro bulblet formation. Plant Cell. Tissue Organ Cult. 1993, 32, 175–183. [Google Scholar] [CrossRef]
- Wiles, G.C. The effect of different photoperiods and temperatures following bulb initiation on bulb development in tropical onion cultivars. Acta Hortic. 1994, 358, 419–427. [Google Scholar] [CrossRef]
- Brewster, J.L. Effects of Photoperiod, Nitrogen Nutrition and Temperature on Inflorescence Initiation and Development in Onion (Allium cepa L.). Ann. Bot. 1983, 51, 429–440. [Google Scholar]
- Okporie, E.O.; Ekpe, I.I. Effect of photoperiod on the growth and bulbing of two tropical onion (Allium cepa L.) varieties. World J. Agric. Sci. 2008, 4, 36–39. [Google Scholar]
- Martínez-García, J.F.; García-Martínez, J.L.; Bou, J.; Prat, S. The Interaction of Gibberellins and Photoperiod in the Control of Potato Tuberization. J. Plant Growth Regul. 2001, 20, 377–386. [Google Scholar] [CrossRef] [PubMed]
- King, R.W.; Moritz, T.; Evans, L.T.; Martin, J.; Andersen, C.H.; Blundell, C.; Kardailsky, I.; Chandler, P.M. Regulation of flowering in the longday grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol. 2006, 141, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Liu, M.S.; Li, J.R.; Guan, C.M.; Zhang, X.S. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol. Biol. 2005, 58, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.J.; Takeuchi, Y.; Kenta, T.; Kume, T.; Bibian, D.; Shimizu, K.K. Mass flowering of the tropical tree Shorea beccariana was preceded by expression changes in flowering and drought-responsive genes. Mol. Ecol. 2013, 22, 4767–4782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. Bioessays 2004, 26, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Ma, X.; Guo, H.; Sukiran, N.L.; Guo, B.; Assmann, S.M.; Ma, H. Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 2013, 25, 3785–3807. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; Colasanti, J.; Rothstein, S.J. The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 2011, 62, 3727–3735. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [PubMed]
- NCBI CDD Database. Available online: http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml (accessed on 14 March 2015).
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- SMART Database. Available online: http://smart.embl-heidelberg.de/ (accessed on 14 March 2015).
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [PubMed]
- CLUSTALX. Available online: http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html (accessed on 9 January 2015).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- ProtParam tool. Available online: http://web.expasy.org/protparam/ (accessed on 30 August 2015).
- EMBOSS. Available online: http://emboss.sourceforge.net/apps/ (accessed on 30 August 2015).
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Plant materials and cDNA samples are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manoharan, R.K.; Han, J.S.H.; Vijayakumar, H.; Subramani, B.; Thamilarasan, S.K.; Park, J.-I.; Nou, I.-S. Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa. Molecules 2016, 21, 217. https://doi.org/10.3390/molecules21020217
Manoharan RK, Han JSH, Vijayakumar H, Subramani B, Thamilarasan SK, Park J-I, Nou I-S. Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa. Molecules. 2016; 21(2):217. https://doi.org/10.3390/molecules21020217
Chicago/Turabian StyleManoharan, Ranjith Kumar, Jeong Suk Hyeon Han, Harshavardhanan Vijayakumar, Boopathi Subramani, Senthil Kumar Thamilarasan, Jong-In Park, and Ill-Sup Nou. 2016. "Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa" Molecules 21, no. 2: 217. https://doi.org/10.3390/molecules21020217
APA StyleManoharan, R. K., Han, J. S. H., Vijayakumar, H., Subramani, B., Thamilarasan, S. K., Park, J.-I., & Nou, I.-S. (2016). Molecular and Functional Characterization of FLOWERING LOCUS T Homologs in Allium cepa. Molecules, 21(2), 217. https://doi.org/10.3390/molecules21020217