Genetic and Epigenetic Approaches for the Possible Detection of Adulteration and Auto-Adulteration in Saffron (Crocus sativus L.) Spice
Abstract
:1. Introduction
2. Results and Discussion
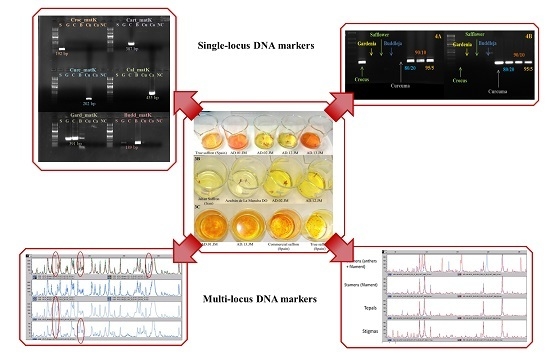
2.1. DNA Extraction and PCR Amplificability
2.2. Primer Design and Evaluation
2.3. AFLP and MS-AFLP Analysis of the Different Saffron Flower Parts
3. Experimental Section
3.1. Sample Set
3.2. DNA Extraction and PCR Amplificability
3.3. Marker Development and Validation
3.4. AFLP and MS-AFLP Analysis of the Saffron Flower Parts
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fernández, J.A. Biology, biotechnology and biomedicine of saffron. Recent. Res. Dev. Plant Sci 2004, 2, 127–159. [Google Scholar]
- Kyriakoudi, A.; Ordoudi, S.A.; Roldán-Medina, M.; Tsimidou, M.Z. Saffron, A Functional Spice. Austin J. Nutr. Food Sci. 2015, 3. [Google Scholar]
- Torelli, A.; Marieschi, M.; Bruni, R. Authentication of saffron (Crocus sativus L.) in different processed, retail products by means of SCAR markers. Food Control 2014, 36, 126–131. [Google Scholar] [CrossRef]
- Food Fraud Database. Available online: https://www.foodfraud.org (accessed on 24 September 2015).
- Europe Saffron White Book. Available online: www.europeansaffron.eu/archivos/White%20book%20english.pdf (accessed on 29 April 2015).
- Petrakis, E.A.; Cagliani, L.R.; Polissiou, M.G.; Consonni, R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by 1H-NMR metabolite fingerprinting. Food Chem. 2015, 173, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Babaei, S.; Talebi, M.; Bahar, M. Developing an SCAR and ITS reliable multiplex PCR-based assay for safflower adulterant detection in saffron samples. Food Control 2014, 35, 323–328. [Google Scholar] [CrossRef]
- Marieschi, M.; Torelli, A.; Bruni, R. Quality control of saffron (Crocus sativus L.): Development of SCAR markers for the detection of plant adulterants used as bulking agents. J. Agric. Food. Chem. 2012, 60, 10998–11004. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Cao, L.; Yuan, Y.; Chen, M.; Jin, Y.; Huang, L. Barcoding melting curve analysis for rapid, sensitive, and discriminating authentication of saffron (Crocus sativus L.) from its adulterants. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hsieh, T.F. Heritable epigenetic variation and its potential applications for crop improvement. Plant Breed Biotechnol. 2013, 1, 307–319. [Google Scholar] [CrossRef]
- Tamari, F.; Hinkley, C.S.; Ramprashad, N. A comparison of DNA extraction methods using Petunia hybrid Tissues. J. Biomol. Tech. 2013, 24, 113–118. [Google Scholar] [PubMed]
- Yang, Z.H.; Xiao, Y.; Zeng, G.M.; Xu, Z.Y.; Liu, Y.S. Comparison of methods for total community DNA extraction and purification from compost. Appl. Microbiol. Biotechnol. 2007, 74, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Foroni, C.; Corradi, M.; Bongiorni, C.; Cattapan, F.; Fogher, C. DNA extraction from olive oil and its use in the identification of the production cultivar. Food Chem. 2003, 83, 127–134. [Google Scholar] [CrossRef]
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; DeMattia, F. DNA barcoding for minor crops and food traceability. Adv. Agric. 2014, 2014. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [PubMed]
- Siracusa, L.; Gresta, F.; Avola, G.; Albertini, E.; Raggi, L.; Marconi, G.; Lombardo, G.M.; Ruberto, G. Agronomic, chemical and genetic variability of saffron (Crocus sativus L.) of different origin by LC-UV-vis-DAD and AFLP analyses. Genet. Resour. Crop. Evol. 2013, 60, 711–721. [Google Scholar] [CrossRef]
- Alsayied, N.F.; Fernández, J.A.; Schwarzacher, T.; Heslop-Harrison, J.S. Diversity and relationships of Crocus sativus and its relatives analysed by inter-retroelement amplified polymorphism (IRAP). Ann. Bot. 2015, 116, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Colli, L.; Sánchez, R.A.; Santaella, M.; Pascual, M.D.L.M.; Santana, O.; Roldán, M.; Fernández, J.A. AFLP and MS-AFLP analysis of the variation within saffron crocus (Crocus sativus L.) germplasm. PLoS ONE 2015, 10, e0123434. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.A.; Santana, O.; Guardiola, J.L.; Molina, R.V.; Heslop-Harrison, P.; Borbely, G.; Branca, F.; Argento, S.; Maloupa, E.; Talou, T.; et al. The world saffron and Crocus collection: Strategies for establishment, management, characterisation and utilisation. Genet. Resour. Crop. Ev. 2011, 58, 125–137. [Google Scholar] [CrossRef]
- Bigliazzi, J.; Scali, M.; Paolucci, E.; Cresti, M.; Vignani, R. DNA extracted with optimised protocols can be genotyped to reconstruct the varietal composition of monovarietal wines. Am. J. Enol. Vitic. 2012, 63, 568–573. [Google Scholar] [CrossRef]
- Demeke, T.; Jenkins, G.R. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 2014, 396, 1977–1990. [Google Scholar] [CrossRef] [PubMed]
- Fazekas, A.J.; Kuzmina, M.L.; Newmaster, S.G.; Hollingsworth, P.M. DNA barcoding methods for land plants. In DNA Barcodes: Methods and Protocols, Methods in Molecular Biology, 1st ed.; Kress, W.J., Erickson, D.L., Eds.; WILEY-VCH Verlag GmbH & Co. KGaA. Publisher: Weinheim, Germany, 2012; Volume 858, pp. 223–252. [Google Scholar]
- Koonjul, P.K.; Brandt, W.F.; Farrant, J.M.; Lindsey, G.G. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 1999, 27, 915–916. [Google Scholar] [CrossRef] [PubMed]
- Busconi, M.; Zacconi, C.; Scolari, G. Bacterial ecology of PDO Coppa and Pancetta Piacentina at the end of ripening and after MAP storage of sliced product. Int. J. Food. Microbiol. 2014, 172, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sample Availability: Crocus sativus accessions are available at the World Saffron and Crocus Collection located at the Bank of Plant Germplasm of Cuenca (Cuenca, Spain). Curcuma, safflower, calendula, buddleia, gardenia, gardenia liquid extracts, gardenia solid extracts and buddleia solid extracts are available at the on-line market. Commercial adulterated saffron is not available from the authors.







| Name a | Description | Origin | DNA Extraction b |
|---|---|---|---|
| AD.01.JM | Saffron (C. sativus) powder adulterated | France | - |
| AD.02.JM | Saffron stigmas adulterated | France | - |
| AD.03.JM | Gardenia (G. jasminoides) extract (liquid) | France | n.v. |
| AD.04.POL | Safflower (C. tinctorius) petals | Greece | + |
| AD.05.POL | Gardenia fruit extract (powder) | Greece | n.v. |
| AD.06.POL | Calendula (C. officinalis) petals | Greece | + |
| AD.07.POL | Saffron stamens | Greece | -- |
| AD.08.POL | Curcuma (C longa) rhizome powder | Greece | -- |
| AD.09.POL | Buddleia (B. officinalis) powder extract | Greece | n.v. |
| AD.10.POL | Buddleia powder extract | Greece | n.v. |
| AD.11.JM | Curcuma powder | France | -- |
| AD.12.JM | Saffron stigmas adulterated | France | - |
| AD.13.JM | Saffron powder adulterated | France | - |
| AD.14.JM | Gardenia extract (liquid) | France | n.v. |
| AD.15.BM | Curcuma powder | Italy | -- |
| AD.16.BM | Saffron powder Commercial A | Italy | - |
| AD.17.BM | Saffron powder Commercial B | Italy | - |
| AD.18.BM | Saffron leaves | Italy | + |
| AD.19.BM | Safflower leaves | Italy | + |
| AD.20.BM | Gardenia leaves | Italy | + |
| AD.21.BM | Buddleia leaves | Italy | + |
| AD.22.JM | Gardenia fruits | France | n.v. |
| Primer Name a | Sequence | Annealing Temperature | Amplicon Size (bp) | Typology |
|---|---|---|---|---|
| matK-KIM1R | ACCCAGTCCATCTGGAAATCTTGGTTC | 58 °C | Variable, circa 900 | Universal |
| matK-KIM3F | CGTACAGTACTTTTGTGTTTACGAG | |||
| rbcL-F | ACCACAAACAGAGACTAAAGC | 52 °C | Variable circa 600 | |
| rbcL-R | GTAAAATCAAGTCCACCRCG | |||
| ITS-S2F | ATGCGATACTTGGTGTGAAT | 52 °C | Variable circa 400 | |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| Gard_matK_Fw | TGGGATACTCTTATTGATAG | 55 °C | 391 | Primers developed in the present study |
| Gard_matK_Rev | CCGGGTGAAACCAAATAC | |||
| Budd_matK_Fw | GAACGTCTTTGTTAAGGTTAAG | 58 °C | 189 | |
| Budd_matK_Rev | CTTGGATGAAACCAAAGCGA | |||
| Curc_matK_Fw | GTAAAAATAGAACATCTTGGAG | 56 °C | 202 | |
| Curc_matK_Rev | ATATGGTTGAGACCAAAAATG | |||
| Cart_matK_Fw | TGTATGTGAATATGAATCTGGC | 54 °C | 387 | |
| Cart_matK_Rev | CCATTGAACGCTTTACCGCG | |||
| Croc_matK_Fw | ATCTTATAATAGTATGTTGTGAT | 54 °C | 192 | |
| Croc_matK_Rev | TGTATGATTGATACCAAAAGT | |||
| Cal_matK_Fw | CATACTCTGGGCCACAAC | 53 °C | 435 | |
| Cal_matK_Rev | GAGGAAGCCGTATTCATATT |
| Sample | Extraction Methods | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GeneElute Plant | Plant DNA Purification | DNeasy Plant | |||||||
| matK | rbcL | ITS | matK | rbcL | ITS | matK | rbcL | ITS | |
| >900 a | >600 | >400 | >900 | >600 | >400 | >900 | >600 | >400 | |
| AD.01.JM | + b | + | + | / | / | / | / | / | / |
| AD.02.JM | + | + | + | / | / | / | / | / | / |
| AD.03.JM | / | / | / | / | / | / | / | / | / |
| AD.04.POL | + | + | + | + | + | + | + | + | + |
| AD.05.POL | / | / | / | / | / | / | / | / | / |
| AD.06.POL | + | + | + | + | + | + | + | + | + |
| AD.07.POL | + | + | + | / | / | / | +/− | +/− | +/− |
| AD.08.POL | + | + | + | / | / | / | / | / | / |
| AD.09.POL | / | / | / | / | / | / | / | / | / |
| AD.10.POL | / | / | / | / | / | / | / | / | / |
| AD.11.JM | + | + | + | / | / | / | / | / | / |
| AD.12.JM | + | + | + | / | / | / | / | / | / |
| AD.13.JM | + | + | + | / | / | / | / | / | / |
| AD.14.JM | / | / | / | / | / | / | / | / | / |
| AD.15.BM | + | + | + | / | / | / | / | / | / |
| AD.16.BM | + | + | + | / | / | / | / | / | / |
| AD.17.BM | + | + | + | / | / | / | / | / | / |
| AD.18.BM | + | + | + | + | + | + | + | + | + |
| AD.19.BM | + | + | + | + | + | + | + | + | + |
| AD.20.BM | + | + | + | + | + | + | + | + | + |
| AD.21.BM | + | + | + | + | + | + | + | + | + |
| AD.22.JM | + | + | + | / | / | / | / | / | / |
| rbcL | Crocus sativus | Gardenia jasminoides | Buddleja officinalis | Curcuma longa | Carthamus tinctorius | Calendula officinalis | matK | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Id% | gap% | Id% | gap% | Id% | gap% | Id% | gap% | Id% | gap% | Id% | gap% | ||
| Crocus sativus | / | / | 73 | 3 | 74 | 2 | 80 | 2 | 75 | 2 | 74 | 2 | Crocus sativus |
| Gardenia jasminoides | 91 | 0 | / | / | 74 | 2 | 76 | 1 | 83 | 1 | 83 | 1 | Gardenia jasminoides |
| Buddleja officinalis | 90 | 0 | 95 | 0 | / | / | 76 | 1 | 85 | 1 | 82 | 1 | Buddleja officinalis |
| Curcuma longa | 94 | 0 | 90 | 0 | 90 | 1 | / | / | 76 | 1 | 74 | 2 | Curcuma longa |
| Carthamus tinctorius | 89 | 0 | 95 | 0 | 94 | 0 | 90 | 0 | / | / | 93 | 0 | Carthamus tinctorius |
| Calendula officinalis | 89 | 0 | 94 | 0 | 94 | 0 | 90 | 0 | 98 | 0 | / | / | Calendula officinalis |
| matK Markers | Saffron | Safflower | Curcuma | Buddleia | Gardenia | Calendula |
|---|---|---|---|---|---|---|
| Croc_matK | + | |||||
| Cart_matK | + | |||||
| Curc_matK | + | |||||
| Gard_matK | + | + | + | |||
| Budd_matK | + | + | ||||
| Cal_matK | + |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soffritti, G.; Busconi, M.; Sánchez, R.A.; Thiercelin, J.-M.; Polissiou, M.; Roldán, M.; Fernández, J.A. Genetic and Epigenetic Approaches for the Possible Detection of Adulteration and Auto-Adulteration in Saffron (Crocus sativus L.) Spice. Molecules 2016, 21, 343. https://doi.org/10.3390/molecules21030343
Soffritti G, Busconi M, Sánchez RA, Thiercelin J-M, Polissiou M, Roldán M, Fernández JA. Genetic and Epigenetic Approaches for the Possible Detection of Adulteration and Auto-Adulteration in Saffron (Crocus sativus L.) Spice. Molecules. 2016; 21(3):343. https://doi.org/10.3390/molecules21030343
Chicago/Turabian StyleSoffritti, Giovanna, Matteo Busconi, Rosa Ana Sánchez, Jean-Marie Thiercelin, Moschos Polissiou, Marta Roldán, and José Antonio Fernández. 2016. "Genetic and Epigenetic Approaches for the Possible Detection of Adulteration and Auto-Adulteration in Saffron (Crocus sativus L.) Spice" Molecules 21, no. 3: 343. https://doi.org/10.3390/molecules21030343
APA StyleSoffritti, G., Busconi, M., Sánchez, R. A., Thiercelin, J.-M., Polissiou, M., Roldán, M., & Fernández, J. A. (2016). Genetic and Epigenetic Approaches for the Possible Detection of Adulteration and Auto-Adulteration in Saffron (Crocus sativus L.) Spice. Molecules, 21(3), 343. https://doi.org/10.3390/molecules21030343