An Approach to Characterizing the Complicated Sequential Metabolism of Salidroside in Rats
Abstract
:1. Introduction
2. Results and Discussion
2.1. Stability of Salidroside in Artificial Gastric Juice and Intestinal Juice
2.2. Fragmentation of Salidroside Standard
2.3. Identification of Salidroside Metabolites in Different Plasma Samples
2.3.1. Parent Compound M0
2.3.2. Metabolite M1
2.3.3. Metabolite M2
2.3.4. Metabolite M3
2.3.5. Metabolite M4
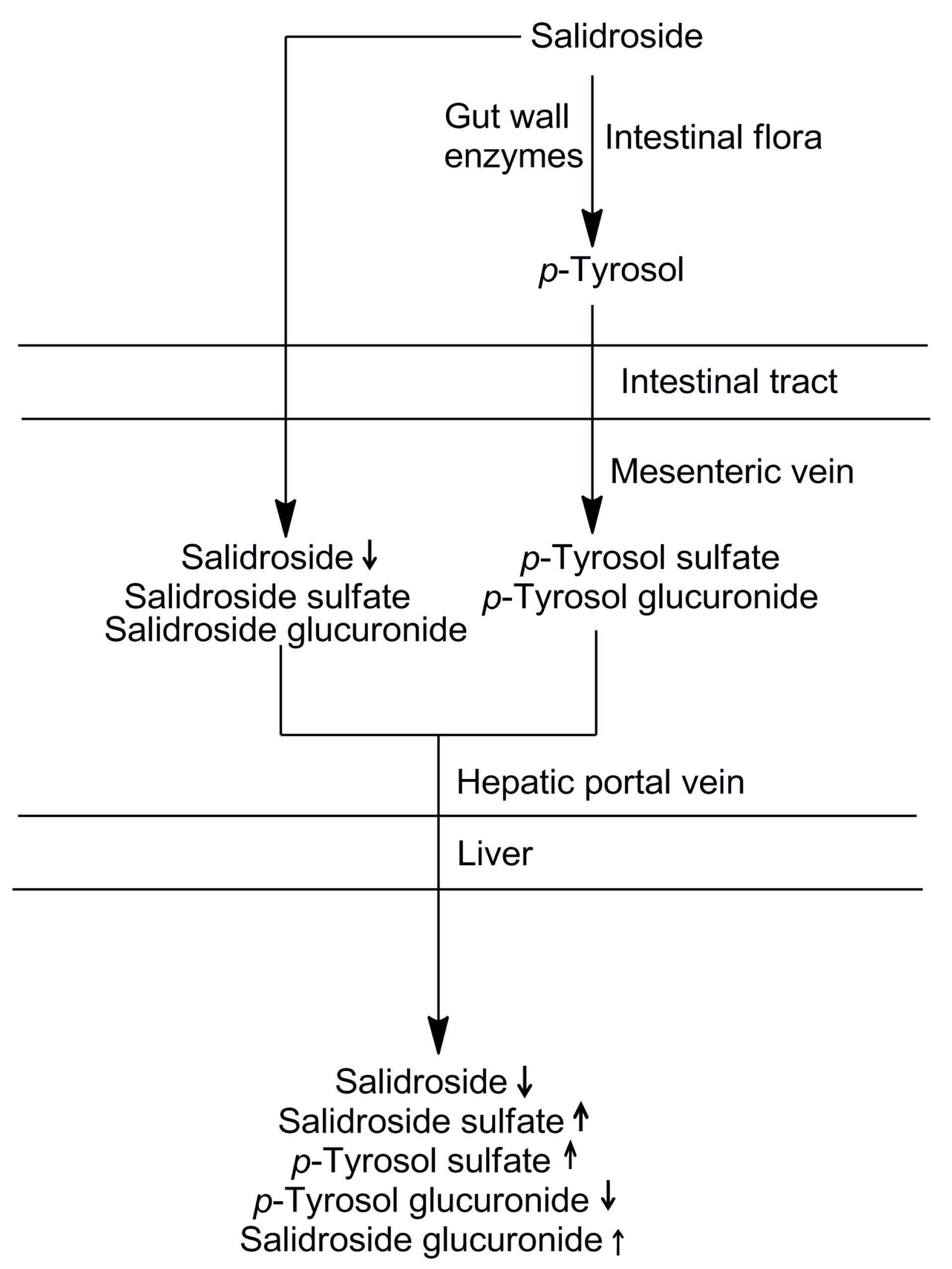
2.4. The Sequential Process of Salidroside in the Digestive System
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Preparation Perfusion Solution of Salidroside
3.3. Animals
3.4. Stability in Digestive Juice with Enzymes
3.5. Intestinal Flora Metabolism Method
3.6. Gut Wall Metabolism Method
3.7. Hepatic Metabolism Method
3.8. HPLC Analysis
3.9. LC/MS Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, S.; Shen, Y.; Liu, J.; Ding, F. Involvement of ERK1/2 pathway in neuroprotection by salidroside against hydrogen peroxide-induced apoptotic cell death. J. Mol. Neurosci. 2010, 40, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, A.; Hou, R.; Zhang, J.; Jia, X.; Jiang, W.; Chen, J. Salidroside protects cardiomyocyte against hypoxia-induced death: A HIF-1α-activated and VEGF-mediated pathway. Eur. J. Pharmacol. 2009, 607, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dong, C.; Huai, L.; Bende, T.; Li, S.; Ying, W. Anti-fatigue effects of salidroside in mice. J. Med. Coll. PLA 2008, 23, 88–93. [Google Scholar] [CrossRef]
- Liu, S.; Yu, X.; Hu, B.; Zou, Y.; Li, J.; Bo, L.; Deng, X. Salidroside rescued mice from experimental sepsis through anti-inflammatory and anti-apoptosis effects. J. Surg. Res. 2015, 195, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Díaz, L.A.; Abad, M.M.; Fernández, M.L.; Recuero, C.C.; Villaescusa, C.L.; Silván, S.A.; Bermejo, B.P. Lignan and phenylpropanoid glycosides from Phillyrea latifolia and their in vitro anti-inflammatory activity. Planta. Med. 2001, 67, 219–223. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Sun, Y.; Lin, X.; Chen, B.; Tan, C.; Cao, G.; Wang, Z. Protective effects of salidroside on hydrogen peroxide-induced apoptosis in SH-SY5Y human neuroblastoma cells. Eur. J. Pharmacol. 2007, 564, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Lin, S.; Yu, D.; Qiu, S.; Zhang, X.; Mei, R. A preliminary study: The anti-proliferation effect of salidroside on different human cancer cell lines. Cell. Biol. Toxicol. 2010, 26, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Chen, Y.; Xu, B.; Ren, K.; He, Z.; He, L.; Lei, Y.; Fan, C.; Song, X. Development of lipid-shell and polymer core nanoparticles with water-soluble salidroside for anti-cancer therapy. Int. J. Mol. Sci. 2014, 15, 3373–3388. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Lu, A.; Zhang, K.; Li, J. Anticancer effect of salidroside on A549 lung cancer cells through inhibition of oxidative stress and phospho-p38 expression. Oncol. Lett. 2014, 7, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shi, Y.; Wu, D.; Ji, Y.; Wang, X.; Chen, H.; Wu, S.; Huang, D.; Jiang, W. Salidroside protects against hydrogen peroxide-induced injury in cardiac H9c2 cells via PI3K-Akt dependent pathway. DNA. Cell. Biol. 2011, 30, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Adaptogen, A.P.P. Rhodiola rosea: A possible plant adaptogen. Altern. Med. Rev. 2001, 6, 293–302. [Google Scholar]
- Nan, J.; Jiang, Y.; Park, E.J.; Ko, G.; Kim, Y.C.; Sohn, D.H. Protective effect of Rhodiola sachalinensis extract on carbon tetrachloride-induced liver injury in rats. J. Ethnopharmacol. 2003, 84, 143–148. [Google Scholar] [CrossRef]
- Zovko Koncic, M.; Tomczyk, M. New insights into dietary supplements used in sport: active substances, pharmacological and side effects. Curr. Drug. Targets. 2013, 14, 1079–1092. [Google Scholar] [CrossRef]
- Skopińska-Rózewska, E.; Malinowski, M.; Wasiutyński, A.; Sommer, E.; Furmanowa, M.; Mazurkiewicz, M.; Siwicki, A. The influence of Rhodiola quadrifida 50% hydro-alcoholic extract and salidroside on tumor-induced angiogenesis in mice. Pol. J. Vet. Sci. 2007, 11, 97–104. [Google Scholar]
- Fan, M.; Xu, S.; Xia, S.; Zhang, X. Preparation of salidroside nano-liposomes by ethanol injection method and in vitro release study. Eur. Food. Res. Technol. 2008, 227, 167–174. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Y.; Zhou, J.; Sun, X.; Wang, S. The in vitro and in vivo antiviral effects of salidroside from Rhodiola rosea L. against coxsackievirus B3. Phytomedicine 2009, 16, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Li, Y.; Mao, X.; Zhang, X.; Guan, J.; Song, A.; Yin, R. Metabolic profile of salidroside in rats using high-performance liquid chromatography combined with Fourier transform ion cyclotron resonance mass spectrometry. Anal. Bioanal. Chem. 2015, 408, 1975–1981. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Wang, Z.; Liu, Y.; Wu, Y.; Han, X.; Zheng, J.; Yan, X.; Wang, Y. Metabolite Profile of Salidroside in Rats by Ultraperformance Liquid Chromatography Coupled with Quadrupole Time-of-Flight Mass Spectrometry and High-Performance Liquid Chromatography Coupled with Quadrupole-Linear Ion Trap Mass Spectrometry. J. Agr. Food. Chem. 2015, 63, 8999–9005. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Zhu, M.; Han, X.; Sui, D.; Wang, Y.; Yang, Q. The metabolism of salidroside to its aglycone p-tyrosol in rats following the administration of salidroside. PLoS ONE 2014, 9, e103648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, H.; Liu, Y.; Dong, H.; Lv, B.; Fang, M.; Zhao, H. Metabolic routes along digestive system of licorice: multicomponent sequential metabolism method in rat. Biomed. Chromatogr. 2015, 30, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Xin, S.; Han, L.; Zuo, R.; Li, Y.; Gong, M.; Wei, X.; Zhou, Y.; He, J.; Wang, H. Comparative metabolism of four limonoids in human liver microsomes using ultra-high-performance liquid chromatography coupled with high-resolution LTQ-Orbitrap mass spectrometry. Rapid. Commun. Mass. Spectrom. 2015, 29, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, W.; Han, F.; Chen, Y. LC/MS/MS for identification of in vivo and in vitro metabolites of jatrorrhizine. Biomed. Chromatogr. 2008, 22, 1360–1367. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Liu, Y.; Zhao, B.; Tang, M.; Dong, H.; Zhang, L.; Lv, B.; Wei, L. Ex vivo and in situ approaches used to study intestinal absorption. J. Pharmacol. Toxicol. 2013, 68, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.


| Metabolite | tR/min | Found (Da) | Calculated (Da) | Error (ppm) | Fragment ion m/z (Da) | Formula | Metabolite Description | Relative Percentage Area in Different Blood Samples 1 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GWM 2 | WOIF | WIIF | HM | ||||||||
| M0 | 11.17 | 299.1126 | 299.1125 | 0.4 | 179.0556 | C14H20O7 | Parent | 94.26% | 99.59% | 43.85% | 22.93% |
| 119.0348 | |||||||||||
| 161.0451 | |||||||||||
| 143.0347 | |||||||||||
| 131.0346 | |||||||||||
| 113.0242 | |||||||||||
| M1 | 8.32 | 313.0932 | 313.0918 | 4.5 | 175.0254 | C14H18O8 | Deglycosylation + Glucuronidation | 2.17% | 0.38% | 15.67% | 7.43% |
| 113.0249 | |||||||||||
| 295.0826 | |||||||||||
| M2 | 8.93 | 475.1464 | 475.1446 | 3.8 | 457.1329 | C20H28O13 | Glucuronidation | 0.66% | 0.04% | 3.52% | 16.42% |
| 175.0242 | |||||||||||
| 299.1121 | |||||||||||
| M3 | 10.80 | 379.0691 | 379.0693 | −0.6 | 217.0170 | C14H20O10S | Sulfation | 2.91% | ND | 7.00% | 11.00% |
| 299.1123 | |||||||||||
| 137.0605 | |||||||||||
| 119.0500 | |||||||||||
| M4 | 11.61 | 217.0173 | 217.0165 | 3.4 | 137.0610 | C8H9O5S | Deglycosylation + Sulfation | ND 3 | ND | 29.96% | 42.23% |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Z.; Ma, X.; Liu, Y.; Lu, L.; Yang, R.; Yu, G.; Sun, M.; Xin, S.; Tian, S.; Chen, X.; et al. An Approach to Characterizing the Complicated Sequential Metabolism of Salidroside in Rats. Molecules 2016, 21, 706. https://doi.org/10.3390/molecules21060706
Luo Z, Ma X, Liu Y, Lu L, Yang R, Yu G, Sun M, Xin S, Tian S, Chen X, et al. An Approach to Characterizing the Complicated Sequential Metabolism of Salidroside in Rats. Molecules. 2016; 21(6):706. https://doi.org/10.3390/molecules21060706
Chicago/Turabian StyleLuo, Zhiqiang, Xiaoyun Ma, Yang Liu, Lina Lu, Ruirui Yang, Guohua Yu, Mohan Sun, Shaokun Xin, Simin Tian, Xinjing Chen, and et al. 2016. "An Approach to Characterizing the Complicated Sequential Metabolism of Salidroside in Rats" Molecules 21, no. 6: 706. https://doi.org/10.3390/molecules21060706





