Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes
Abstract
:1. Introduction
2. Results
2.1. Shikonin Inhibits the Proliferation of MCF-7 Cells in Time- and Dose-Dependent Manners
2.2. Shikonin Inhibits Exosome Release in MCF-7 Cells
2.3. Shikonin Inhibits MCF-7 Cell Proliferation by Suppressing Its Exosome Release
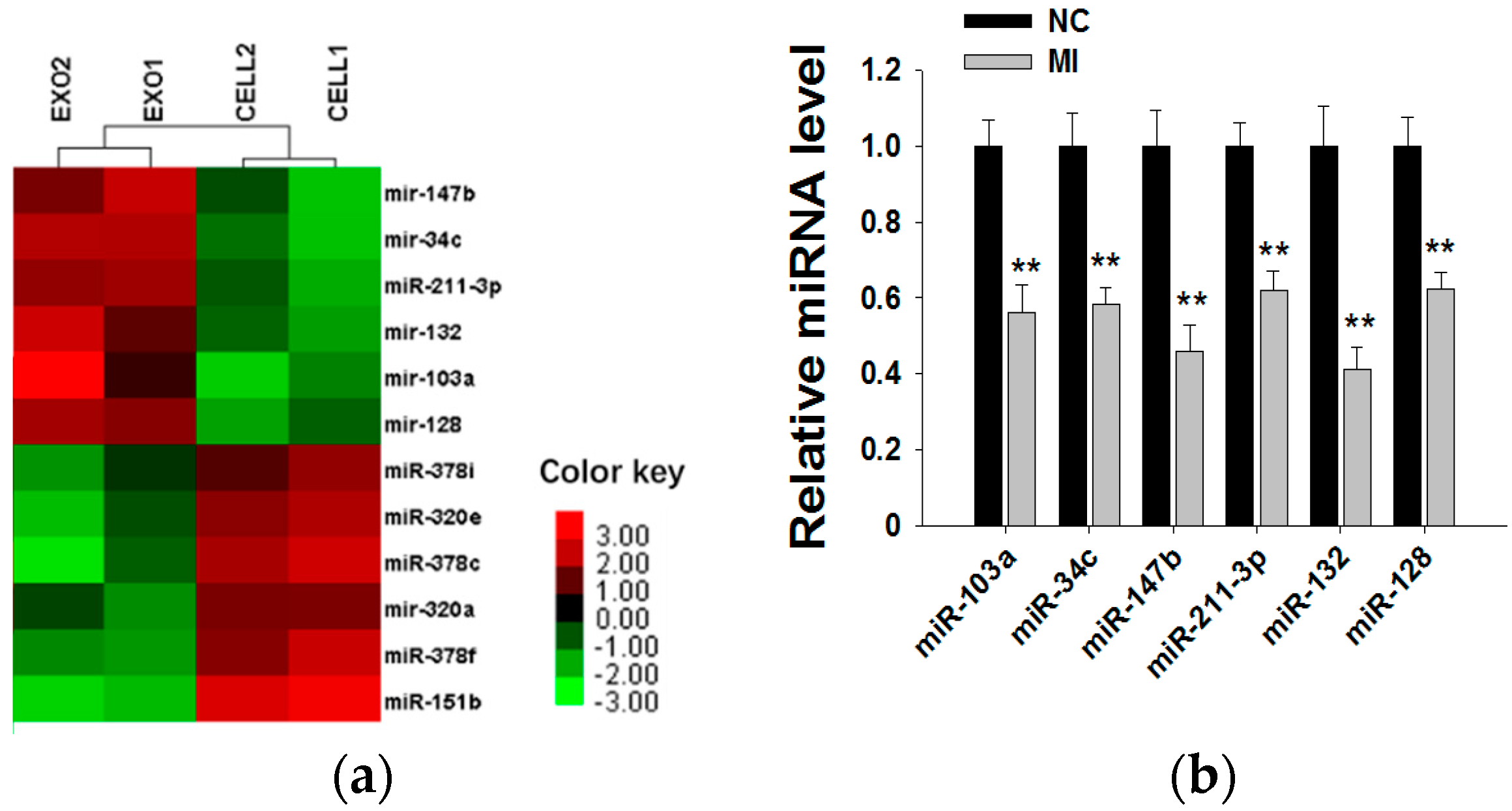
2.4. Shikonin Decreases Exosomal miR-128 to Inhibit MCF-7 Cell Proliferation
2.5. miR-128 Promotes MCF-7 Cell Proliferation by Targeting the Bax Gene
3. Discussion
4. Materials and Methods
4.1. Cells and Reagents
4.2. Cell Proliferation Assay
4.3. Isolation of Exosomes
4.4. Transmission Electron Microscopy Assay
4.5. Nanoparticle Tracking Analysis (NTA)
4.6. Immunofluorescence
4.7. Transfection of Cells with miRNA Inhibitor and Mimic
4.8. RNA Isolation and qRT-PCR of mRNA and Mature miRNAs
4.9. Immunoblotting
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bertucci, F.; Birnbaum, D. Reasons for breast cancer heterogeneity. J. Biol. 2008, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Kuller, L.H. The etiology of breast cancer—From epidemiology to prevention. Public Health Rev. 1995, 23, 157–213. [Google Scholar] [PubMed]
- Malki, A.; Mohsen, M.; Aziz, H.; Rizk, O.; Shaban, O.; El-Sayed, M.; Sherif, Z.A.; Ashour, H. New 3-Cyano-2-Substituted Pyridines Induce Apoptosis in MCF 7 Breast Cancer Cells. Molecules 2016, 21, 230. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Bishayee, A. Mechanism of Breast Cancer Preventive Action of Pomegranate: Disruption of Estrogen Receptor and Wnt/beta-Catenin Signaling Pathways. Molecules 2015, 20, 22315–22328. [Google Scholar] [CrossRef] [PubMed]
- Sapio, L.; Sorvillo, L.; Illiano, M.; Chiosi, E.; Spina, A.; Naviglio, S. Inorganic Phosphate Prevents Erk1/2 and Stat3 Activation and Improves Sensitivity to Doxorubicin of MDA-MB-231 Breast Cancer Cells. Molecules 2015, 20, 15910–15928. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.Y.; Kim, C.F.; Leung, K.N.; Fung, K.P.; Tse, T.F.; Chan, H.; Lau, C.B. Differential anti-tumor activity of coriolus versicolor (Yunzhi) extract through p53- and/or Bcl-2-dependent apoptotic pathway in human breast cancer cells. Cancer Biol. Ther. 2005, 4, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Guo, T.; Wu, C.; He, X.; Zhao, M. Effect of shikonin on human breast cancer cells proliferation and apoptosis in vitro. Yakugaku Zasshi 2006, 126, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.R.; Zhang, Y.; Tang, X. Shikonin inhibits the proliferation of human lens epithelial cells by inducing apoptosis through ROS and caspase-dependent pathway. Molecules 2014, 19, 7785–7797. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ma, C.; Yang, L.; Wang, W.; Sui, X.; Zhao, C.; Zu, Y. Optimization of shikonin homogenate extraction from Arnebia euchroma using response surface methodology. Molecules 2013, 18, 466–481. [Google Scholar] [CrossRef] [PubMed]
- Damianakos, H.; Kretschmer, N.; Syklowska-Baranek, K.; Pietrosiuk, A.; Bauer, R.; Chinou, I. Antimicrobial and cytotoxic isohexenylnaphthazarins from Arnebia euchroma (Royle) Jonst. (Boraginaceae) callus and cell suspension culture. Molecules 2012, 17, 14310–14322. [Google Scholar] [CrossRef] [PubMed]
- Andujar, I.; Rios, J.L.; Giner, R.M.; Recio, M.C. Pharmacological properties of shikonin—A review of literature since 2002. Planta Med. 2013, 79, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, V.P.; Assimopoulou, A.N.; Ballis, A.C. Alkannins and shikonins: A new class of wound healing agents. Curr. Med. Chem. 2008, 15, 3248–3267. [Google Scholar] [CrossRef] [PubMed]
- Kourounakis, A.P.; Assimopoulou, A.N.; Papageorgiou, V.P.; Gavalas, A.; Kourounakis, P.N. Alkannin and shikonin: Effect on free radical processes and on inflammation—A preliminary pharmacochemical investigation. Arch. Pharm. 2002, 335, 262–266. [Google Scholar] [CrossRef]
- Masuda, Y.; Nishida, A.; Hori, K.; Hirabayashi, T.; Kajimoto, S.; Nakajo, S.; Kondo, T.; Asaka, M.; Nakaya, K. Beta-hydroxyisovalerylshikonin induces apoptosis in human leukemia cells by inhibiting the activity of a polo-like kinase 1 (PLK1). Oncogene 2003, 22, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, L.; Oppenheim, J.J.; Howard, M.Z. Cellular pharmacology studies of shikonin derivatives. Phytother. Res. PTR 2002, 16, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Shima, G.; Aiuchi, T.; Horie, M.; Hori, K.; Nakajo, S.; Kajimoto, S.; Shibayama-Imazu, T.; Nakaya, K. Involvement of tumor necrosis factor receptor-associated protein 1 (TRAP1) in apoptosis induced by beta-hydroxyisovalerylshikonin. J. Biol. Chem. 2004, 279, 42503–42515. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Pouyssegur, J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 2008, 13, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Lee, J.K.; Jang, E.H.; Jeong, S.Y.; Kim, J.H. Shikonin blocks migration and invasion of human breast cancer cells through inhibition of matrix metalloproteinase-9 activation. Oncol. Rep. 2014, 31, 2827–2833. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Escudier, B.; Angevin, E.; Tursz, T.; Zitvogel, L. Exosomes for cancer immunotherapy. Ann. Oncol. 2004, 15 (Suppl. 4), iv141–iv144. [Google Scholar]
- Wei, Y.; Li, L.; Wang, D.; Zhang, C.Y.; Zen, K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014, 289, 10270–10275. [Google Scholar] [CrossRef]
- Li, L.; Zhu, D.; Huang, L.; Zhang, J.; Bian, Z.; Chen, X.; Liu, Y.; Zhang, C.Y.; Zen, K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS ONE 2012, 7, e46957. [Google Scholar] [CrossRef]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, F.; de Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef] [PubMed]
- Van Balkom, B.W.; de Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T.; et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 2013, 121, 3997–4006. [Google Scholar] [CrossRef] [PubMed]
- Martins, V.R.; Dias, M.S.; Hainaut, P. Tumor-cell-derived microvesicles as carriers of molecular information in cancer. Curr. Opin. Oncol. 2013, 25, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Shao, G.; Lv, X.; Liu, Y.; Fan, Y.; Wu, A.; Hu, H. Downregulation of miRNA-128 sensitises breast cancer cell to chemodrugs by targeting Bax. Cell Biol. Int. 2013, 37, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Dewson, G.; Ma, S.; Frederick, P.; Hockings, C.; Tan, I.; Kratina, T.; Kluck, R.M. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012, 19, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.







© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.-Y.; Zen, K.; Li, L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules 2016, 21, 777. https://doi.org/10.3390/molecules21060777
Wei Y, Li M, Cui S, Wang D, Zhang C-Y, Zen K, Li L. Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes. Molecules. 2016; 21(6):777. https://doi.org/10.3390/molecules21060777
Chicago/Turabian StyleWei, Yao, Mingzhen Li, Shufang Cui, Dong Wang, Chen-Yu Zhang, Ke Zen, and Limin Li. 2016. "Shikonin Inhibits the Proliferation of Human Breast Cancer Cells by Reducing Tumor-Derived Exosomes" Molecules 21, no. 6: 777. https://doi.org/10.3390/molecules21060777




