Precision or Personalized Medicine for Cancer Chemotherapy: Is there a Role for Herbal Medicine
Abstract
:1. Introduction: Current Limitations of Cancer Chemotherapy
2. Precision Medicine vs. Personalized Medicine—Potential Application to Improved Cancer Chemotherapy
2.1. Definition of Precision Medicine and Personalized Medicine
2.2. Examples of Precision Medicine or Personalized Medicine in Cancer Chemotherapy
2.3. Precision Medicine vs. Personalized Medicine—Perspectives in Drug Development and Practice
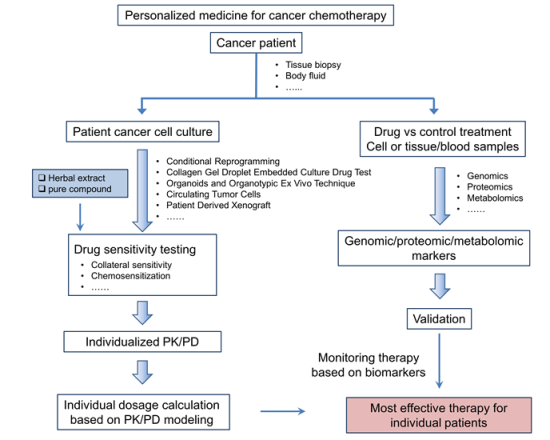
3. Technologies Relevant to Development of Herbal Medicine for Personalized Cancer Chemotherapy
3.1. Biomarker Identification
3.2. Culture of Patient Cancer Cells and Determination of Drug Sensitivity
3.2.1. Collagen Gel Droplet Embedded Culture Drug Test (CD-DST)
3.2.2. Conditional Reprogramming (CR)
3.2.3. Organoids and Organotypic Ex Vivo Technique
3.2.4. Circulating Tumor Cells (CTC)
3.2.5. Patient Derived Xenograft (PDX) Model
3.2.6. Appraisal of the Cell Culture and Sensitivity Techniques for Herbal Product Evaluation
3.3. Pharmacokinetic Approach for Optimal Dosing to Achieve Efficacy and Safety
4. Special Features of Herbal Medicine for Personalized Cancer Chemotherapy—Implications for Future Therapeutic Advancement
4.1. Chemosensitizing Effect (CE)
4.2. Collateral Sensitivity (CS) of Herbs
5. Perspective and Future Direction of Herbal Medicine—Is there a Role for Personalized Medicine in Cancer Chemotherapy
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Coley, H.M. Overcoming multidrug resistance in cancer: An update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control 2003, 10, 159–165. [Google Scholar] [PubMed]
- Wang, Z.; Xie, C.; Huang, Y.; Lam, C.; Chow, M. Overcoming chemotherapy resistance with herbal medicines: Past, present and future perspectives. Phytochem. Rev. 2014, 13, 323–337. [Google Scholar] [CrossRef]
- Rottenberg, S.; Borst, P. Drug resistance in the mouse cancer clinic. Drug Resist. Updates 2012, 15, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G.; Rath, B. A short update on cancer chemoresistance. Wien. Med. Wochenschr. 2014, 164, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Schnipper, L.E.; Davidson, N.E.; Wollins, D.S.; Tyne, C.; Blayney, D.W.; Blum, D.; Dicker, A.P.; Ganz, P.A.; Hoverman, J.R.; Langdon, R. American Society of Clinical Oncology statement: A conceptual framework to assess the value of cancer treatment options. J. Clin. Oncol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Tyagi, A. Traditional Medicine: The Goldmine for Modern Drugs. Adv. Tech. Biol. Med. 2015, 3. [Google Scholar] [CrossRef]
- Roden, D.M.; Tyndale, R.F. Genomic medicine, precision medicine, personalized medicine: What’s in a name? Clin. Pharmacol. Ther. 2013, 94, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Redekop, W.K.; Mladsi, D. The faces of personalized medicine: A framework for understanding its meaning and scope. Value Health 2013, 16, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Wang, W.; Li, A.; Kansal, R.; Chen, Y.; Chen, H.; Li, X. Clinical Next Generation Sequencing for Precision Medicine in Cancer. Curr. Genom. 2015, 16, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Zaneveld, J.; Wang, F.; Wang, X.; Chen, R. Dawn of ocular gene therapy: Implications for molecular diagnosis in retinal disease. Sci. China Life Sci. 2013, 56, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Shi, L.; Hong, H.; Su, Z.; Fuscoe, J.; Ning, B. Studies on abacavir-induced hypersensitivity reaction: A successful example of translation of pharmacogenetics to personalized medicine. Sci. China Life Sci. 2013, 56, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S. Pharmaceutical education in the wake of genomic technologies for drug development and personalized medicine. Eur. J. Pharm. Sci. 2002, 15, 243–250. [Google Scholar] [CrossRef]
- Chen, G.; Shi, T. Next-generation sequencing technologies for personalized medicine: Promising but challenging. Sci. China. Life Sci. 2013, 56, 101. [Google Scholar] [CrossRef] [PubMed]
- Mesri, M. Advances in Proteomic Technologies and Its Contribution to the Field of Cancer. Adv. Med. 2014, 2014, 238045. [Google Scholar] [CrossRef] [PubMed]
- Eckhard, U.; Marino, G.; Butler, G.S.; Overall, C.M. Positional proteomics in the era of the human proteome project on the doorstep of precision medicine. Biochimie 2016, 122, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.-H.; Snyder, M. Omics profiling in precision oncology. Mol. Cell. Proteom. 2016. [Google Scholar] [CrossRef] [PubMed]
- Klement, G.L.; Arkun, K.; Valik, D.; Roffidal, T.; Hashemi, A.; Klement, C.; Carmassi, P.; Rietman, E.; Slaby, O.; Mazanek, P.; et al. Future paradigms for precision oncology. Oncotarget 2016, 19. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Qiu, Y.; Chen, T.; Lin, J.; Chi, Y.; Su, M.; Zhao, A.; Jia, W. An optimized procedure for metabonomic analysis of rat liver tissue using gas chromatography/time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2010, 52, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Fordahl, S.; Cooney, P.; Qiu, Y.; Xie, G.; Jia, W.; Erikson, K.M. Waterborne manganese exposure alters plasma, brain, and liver metabolites accompanied by changes in stereotypic behaviors. Neurotoxicol. Teratol. 2012, 34, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Cortés, J.; Kim, S.-B.; Im, S.-A.; Hegg, R.; Im, Y.-H.; Roman, L.; Pedrini, J.L.; Pienkowski, T.; Knott, A.; et al. Pertuzumab plus Trastuzumab plus Docetaxel for Metastatic Breast Cancer. N. Engl. J. Med. 2012, 366, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, A.; Pasic, M.D.; Yousef, G.M. Proteomics and peptidomics: Moving toward precision medicine in urological malignancies. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wu, F.; Ding, S.; Sun, L.; Liu, Z.; Ding, K.; Lu, J. Label-free quantitative proteomic analysis reveals potential biomarkers and pathways in renal cell carcinoma. Tumor Biol. 2015, 36, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Berman, D.M.; Rimm, D.L.; Shipitsin, M.; Putzi, M.; Nifong, T.P.; Small, C.; Choudhury, S.; Capela, T.; Coupal, L.; et al. Development and Clinical Validation of an in situ Biopsy-Based Multimarker Assay for Risk Stratification in Prostate Cancer. Am. Assoc. Cancer Res. 2015, 21, 2591–2600. [Google Scholar] [CrossRef] [PubMed]
- Shipitsin, M.; Small, C.; Choudhury, S.; Giladi, E.; Friedlander, S.; Nardone, J.; Hussain, S.; Hurley, A.D.; Ernst, C.; Huang, Y.E.; et al. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br. J. Cancer 2014, 111, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Flatley, B.; Wilmott, K.G.; Malone, P.; Cramer, R. MALDI MS profiling of post-DRE urine samples highlights the potential of β-microseminoprotein as a marker for prostatic diseases. Prostate 2014, 74, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Boehm, J.S.; Golub, T.R. An ecosystem of cancer cell line factories to support a cancer dependency map. Nat. Rev. Genet. 2015, 16, 373–374. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Frei, E. The National Cancer Chemotherapy Program. Science 1982, 217, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Venditti, J.M. The National Cancer Institute antitumor drug discovery program, current and future perspectives: A commentary. Cancer Treat. Rep. 1983, 67, 767–772. [Google Scholar] [PubMed]
- Genovese, L.; Zawada, L.; Tosoni, A.; Ferri, A.; Zerbi, P.; Allevi, R.; Nebuloni, M.; Alfano, M. Cellular localization, invasion, and turnover are differently influenced by healthy and tumor-derived extracellular matrix. Tissue Eng. Part A 2014, 20, 2005–2018. [Google Scholar] [CrossRef] [PubMed]
- Schrag, D.; Garewal, H.S.; Burstein, H.J.; Samson, D.J.; Von Hoff, D.D.; Somerfield, M.R. American Society of Clinical Oncology Technology Assessment: Chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2004, 22, 3631–3638. [Google Scholar] [CrossRef] [PubMed]
- Burstein, H.J.; Mangu, P.B.; Somerfield, M.R.; Schrag, D.; Samson, D.; Holt, L.; Zelman, D.; Ajani, J.A. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2011, 29, 3328–3330. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H. Development of a new in vitro chemosensitivity test using collagen gel droplet embedded culture and image analysis for clinical usefulness. Recent Results Cancer Res. Fortschr. Krebsforsch. Progres Dans Les Recherches Sur Le Cancer 2003, 161, 48–61. [Google Scholar]
- Naitoh, H.; Yamamoto, H.; Murata, S.; Kobayashi, H.; Inoue, K.; Tani, T. Stratified phase II trial to establish the usefulness of the collagen gel droplet embedded culture-drug sensitivity test (CD-DST) for advanced gastric cancer. Gastric Cancer 2014, 17, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Higashiyama, M.; Oda, K.; Okami, J.; Maeda, J.; Kodama, K.; Imamura, F.; Minamikawa, K.; Takano, T.; Kobayashi, H. Prediction of chemotherapeutic effect on postoperative recurrence by in vitro anticancer drug sensitivity testing in non-small cell lung cancer patients. Lung Cancer 2010, 68, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Nagai, N.; Minamikawa, K.; Mukai, K.; Hirata, E.; Komatsu, M.; Kobayashi, H. Predicting the chemosensitivity of ovarian and uterine cancers with the collagen gel droplet culture drug-sensitivity test. Anti-Cancer Drugs 2005, 16, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Myers, S.; Wang, J.; Zhou, D.; Woo, J.A.; Kallakury, B.; Ju, A.; Bazylewicz, M.; Carter, Y.M.; Albanese, C.; et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 2012, 367, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Crystal, A.S.; Shaw, A.T.; Sequist, L.V.; Friboulet, L.; Niederst, M.J.; Lockerman, E.L.; Frias, R.L.; Gainor, J.F.; Amzallag, A.; Greninger, P.; et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014, 346, 1480–1486. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Chen, Y. Organoid development in cancer genome discovery. Curr. Opin. Genet. Dev. 2015, 30, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Majumder, B.; Baraneedharan, U.; Thiyagarajan, S.; Radhakrishnan, P.; Narasimhan, H.; Dhandapani, M.; Brijwani, N.; Pinto, D.D.; Prasath, A.; Shanthappa, B.U.; et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.X.; Xue, H.; Lin, D.; Dong, X.; Gout, P.W.; Gao, X.; Pang, J. Subrenal capsule grafting technology in human cancer modeling and translational cancer research. Differentiation 2015, 91, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.L.; Becker, M.A.; Haluska, P.; Samimi, G. Patient-derived xenograft models to improve targeted therapy in epithelial ovarian cancer treatment. Front. Oncol. 2013, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Xue, H.; Wang, Y.; Wu, R.; Watahiki, A.; Dong, X.; Cheng, H.; Wyatt, A.W.; Collins, C.C.; Gout, P.W. Next generation patient-derived prostate cancer xenograft models. Asian J. Androl. 2014, 16, 407. [Google Scholar] [PubMed]
- Van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.; Walters, M.A. Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Kerkhofs, T.M.; Derijks, L.J.; Ettaieb, H.; Den Hartigh, J.; Neef, K.; Gelderblom, H.; Guchelaar, H.-J.; Haak, H.R. Development of a pharmacokinetic model of mitotane: Toward personalized dosing in adrenocortical carcinoma. Ther. Drug Monit. 2015, 37, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Ballesta, A.; Zhou, Q.; Zhang, X.; Lv, H.; Gallo, J. Multiscale Design of Cell-Type–Specific Pharmacokinetic/Pharmacodynamic Models for Personalized Medicine: Application to Temozolomide in Brain Tumors. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Birtwistle, M.; Gallo, J. A General Network Pharmacodynamic Model–Based Design Pipeline for Customized Cancer Therapy Applied to the VEGFR Pathway. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sawchuk, R.J.; Zaske, D.E. Pharmacokinetics of dosing regimens which utilize multiple intravenous infusions: Gentamicin in burn patients. J. Pharmacokinet. Biopharm. 1976, 4, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Platt, D.R.; Matthews, S.J.; Sevka, M.J.; Comer, J.B.; Quintiliani, R.; Cunha, B.A.; Nightingale, C.H.; Chow, M.S. Comparison of four methods of predicting serum gentamicin concentrations in adult patients with impaired renal function. Clin. Pharm. 1982, 1, 361–365. [Google Scholar] [PubMed]
- Burton, M.E.; Chow, M.S.; Platt, D.R.; Day, R.B.; Brater, D.C.; Vasko, M.R. Accuracy of Bayesian and Sawchuk-Zaske dosing methods for gentamicin. Clin. Pharm. 1986, 5, 143–149. [Google Scholar] [PubMed]
- Ling, C.-Q.; Yue, X.-Q.; Ling, C. Three advantages of using traditional Chinese medicine to prevent and treat tumor. J. Integr. Med. 2014, 12, 331–335. [Google Scholar] [CrossRef]
- Pluchino, K.M.; Hall, M.D.; Goldsborough, A.S.; Callaghan, R.; Gottesman, M.M. Collateral sensitivity as a strategy against cancer multidrug resistance. Drug Resist. Updates 2012, 15, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Marshall, T.S.; Kwit, A.D.T.; Miller Jenkins, L.M.; Dulcey, A.E.; Madigan, J.P.; Pluchino, K.M.; Goldsborough, A.S.; Brimacombe, K.R.; Griffiths, G.L.; et al. Inhibition of Glutathione Peroxidase Mediates the Collateral Sensitivity of Multidrug-resistant Cells to Tiopronin. J. Biol. Chem. 2014, 289, 21473–21489. [Google Scholar] [CrossRef] [PubMed]
- Imamovic, L.; Sommer, M.O. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munck, C.; Gumpert, H.K.; Wallin, A.I.; Wang, H.H.; Sommer, M.O. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 2014, 6, 262ra156. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.L.; Hsia, C.R.; Hsu, C.L.; Huang, H.C.; Juan, H.F. Integrating transcriptomics and proteomics to show that tanshinone IIA suppresses cell growth by blocking glucose metabolism in gastric cancer cells. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.; Wang, X.; Xu, N.; Zhang, H.; Xu, H. Application of proteomics to determine the mechanism of action of traditional Chinese medicine remedies. J. Ethnopharmacol. 2014, 155, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, G.T.; Roh, S.H.; Song, J.S.; Kim, H.J.; Hong, S.S.; Kwon, S.W.; Park, J.H. Proteomic analysis of the anti-cancer effect of 20S-ginsenoside Rg3 in human colon cancer cell lines. Biosci. Biotechnol. Biochem. 2009, 73, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Miyachi, H.; Bartsch, H. Pharmacogenomics of a traditional Japanese herbal medicine (Kampo) for cancer therapy. Cancer Genom. Proteom. 2007, 4, 81–91. [Google Scholar]
- Yun, H.; Hou, L.; Song, M.; Wang, Y.; Zakus, D.; Wu, L.; Wang, W. Genomics and Traditional Chinese Medicine: A New Driver for Novel Molecular-Targeted Personalized Medicine? Curr. Pharmacogenomics Pers. Med. 2016, 10, 6. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Natl. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–715. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, X.; Ho, R.L.K.Y.; Lam, C.W.K.; Chow, M.S.S. Precision or Personalized Medicine for Cancer Chemotherapy: Is there a Role for Herbal Medicine. Molecules 2016, 21, 889. https://doi.org/10.3390/molecules21070889
Wang Z, Liu X, Ho RLKY, Lam CWK, Chow MSS. Precision or Personalized Medicine for Cancer Chemotherapy: Is there a Role for Herbal Medicine. Molecules. 2016; 21(7):889. https://doi.org/10.3390/molecules21070889
Chicago/Turabian StyleWang, Zhijun, Xuefeng Liu, Rebecca Lucinda Ka Yan Ho, Christopher Wai Kei Lam, and Moses Sing Sum Chow. 2016. "Precision or Personalized Medicine for Cancer Chemotherapy: Is there a Role for Herbal Medicine" Molecules 21, no. 7: 889. https://doi.org/10.3390/molecules21070889