Potential Application of p-Coumaric Acid on Differentiation of C2C12 Skeletal Muscle and 3T3-L1 Preadipocytes—An in Vitro and in Silico Approach
Abstract
:1. Introduction
2. Results
2.1. Cytotoxicity of CA on C2C12 and 3T3-L1 Cells
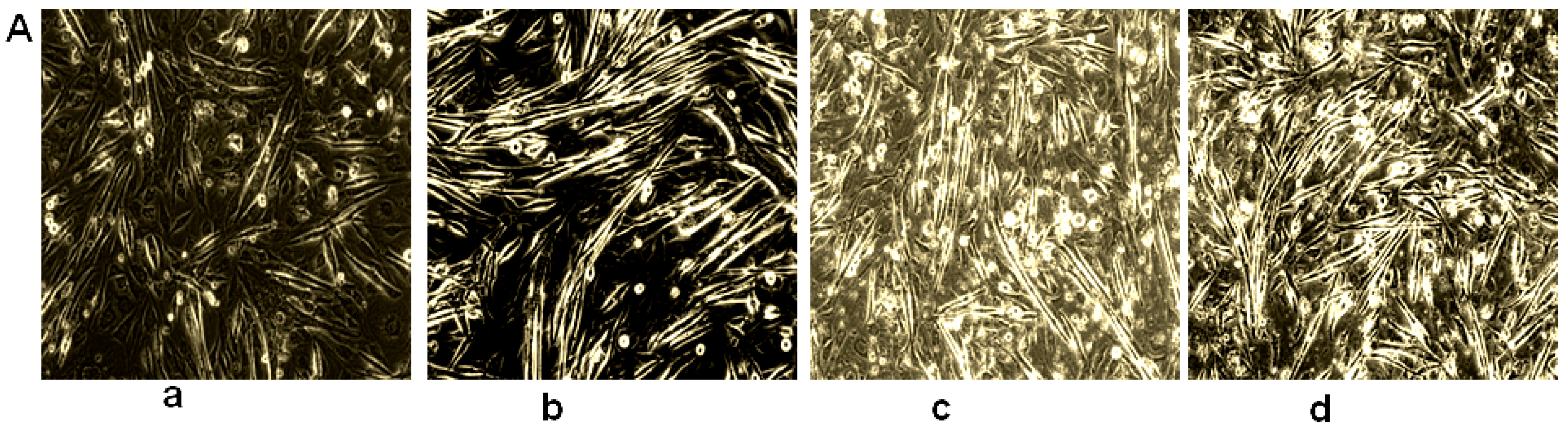
2.2. Coumaric Acid Inhibits Myotube Formation and Early Differentiation Markers in C2C12 Cell
2.3. Coumaric Acid Inhibits Differentiation and Lipid Accumulation in 3T3-L1 Cells
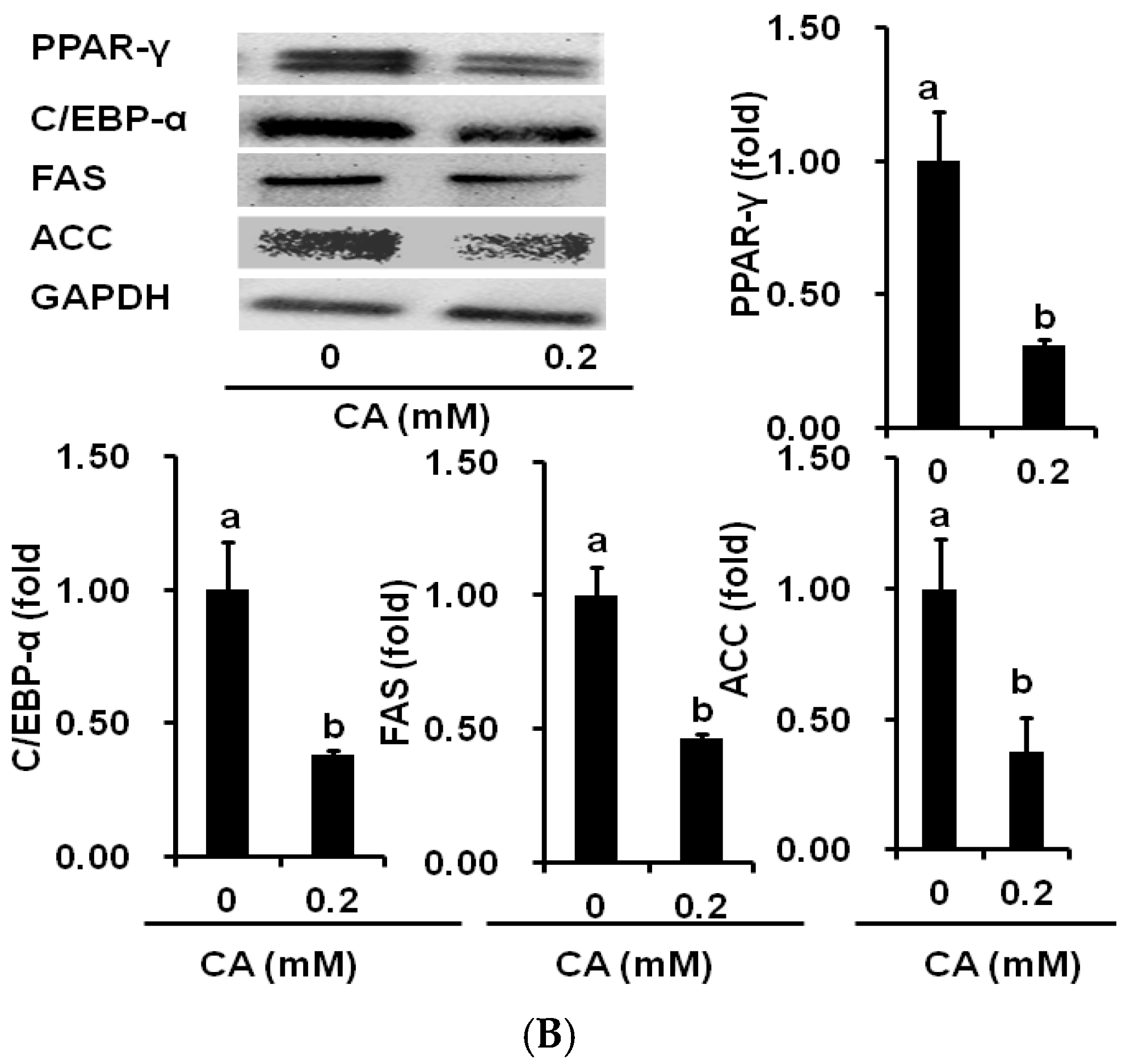
2.4. CA Downregulates Adipogenic and Lipogenic Gene Expression
2.5. Comparative Study between CA, TRZ, and T0070907 on PPAR-γ2 Expression
2.6. Molecular Modelling for CA with PPAR-γ2
3. Discussion
3.1. Coumaric Acid on C2C12 Cell Differentiation
3.2. Coumaric Acid on 3T3-L1 Cell Differentiation
4. Materials and Methods
4.1. Cell Culture and Chemicals
4.2. Cytotoxicity Assay
4.3. Differentiation Induction Protocol for C2C12 Skeletal Muscle
4.4. Differentiation Induction Protocol for 3T3-L1 preadipocytes
4.5. Quantification of Lipid Content by Oil Red O Staining
4.6. Quantification of Adipogenic Gene Expression Using Quantitative RT-PCR
4.7. Protein Extraction and Immunoblotting
4.8. Molecular Modeling
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klip, A.; Pâquet, M.R. Glucose transport and glucose transporters in muscle and their metabolic regulation. Diabetes Care 1990, 13, 228–243. [Google Scholar] [CrossRef] [PubMed]
- Cortright, R.N.; Muoio, D.M.; Dohm, G.L. Skeletal muscle lipid metabolism: A frontier for new insights into fuel homeostasis. J. Nutr. Biochem. 1997, 8, 228–245. [Google Scholar] [CrossRef]
- Doherty, J.T.; Lenhart, K.C.; Cameron, M.V.; Mack, C.P.; Conlon, F.L.; Taylor, J.M. Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1. J. Biol. Chem. 2011, 286, 25903–25921. [Google Scholar] [CrossRef] [PubMed]
- Dedieu, S.; Mazeres, G.; Cottin, P.; Brustis, J.J. Involvement of myogenic regulator factors during fusion in the cell line C2C12. Int. J. Dev. Biol. 2002, 46, 235–241. [Google Scholar]
- Rudnicki, M.A.; Schnegelsberg, P.N.; Stead, R.H.; Braun, T.; Arnold, H.H.; Jaenisch, R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 1993, 75, 1351–1359. [Google Scholar] [CrossRef]
- Kablar, B.; Krastel, K.; Ying, C.; Asakura, A.; Tapscott, S.J.; Rudnicki, M.A. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 1997, 124, 4729–4738. [Google Scholar]
- Ishibashi, J.; Perry, R.L.; Asakura, A.; Rudnicki, M.A. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 2005, 171, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Jiang, M.S.; Lane, M.D. Repressive effect of Sp1 on the C/EBP alpha gene promoter: Role in adipocyte differentiation. Mol. Cell Biol. 1999, 19, 4855–4865. [Google Scholar] [CrossRef]
- Fruhbeck, G.; Gomez-Ambrosi, J.; Muruzabal, F.J.; Burrell, M.A. The adipocyte: A model for integration of endocrine and metabolic signaling in energy metabolism regulation. Am. J. Physiol. Endocrinol. Metab. 2001, 280, 827–847. [Google Scholar]
- Jeon, T.; Hwang, S.G.; Hirai, S.; Matsui, T.; Yano, H.; Kawada, T.; Lim, B.O.; Park, D.K. Red yeast rice extracts suppress adipogenesis by down-regulating adipogenic transcription factors and gene expression in 3T3-L1 cells. Life Sci. 2004, 12, 3195–3203. [Google Scholar] [CrossRef] [PubMed]
- Avram, M.M.; Avram, A.S.; James, W.D. Subcutaneous fat in normal and diseased states: 3. Adipogenesis: From stem cell to fat cell. J. Am. Acad. Dermatol. 2007, 56, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 28, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Christy, R.J.; Kaestner, K.H.; Geiman, D.E.; Lane, M.D. CCAAT/enhancer binding protein gene promoter: Binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 1991, 15, 2593–2597. [Google Scholar] [CrossRef]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar]
- El-Seedi, H.R.; El-Said, A.M.; Khalifa, S.A.; Göransson, U.; Bohlin, L.; Borg-Karlson, A.K.; Veroorte, R. Biosynthesis, natural sources, dietary intake, pharmacokinetic properties, and biological activities of hydroxycinnamic acids. J. Agric. Food Chem. 2012, 60, 10877–10895. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.A.; Kang, S.I.; Shin, H.S.; Kang, S.W.; Kim, J.H.; Ko, H.C.; Kim, S.J. p-Coumaric acid modulates glucose and lipid metabolism via AMP-activated protein kinase in L6 skeletal muscle cells. Biochem. Biophys. Res. Commun. 2013, 432, 553–557. [Google Scholar] [PubMed]
- Hsu, C.L.; Yen, G.C. Effects of flavonoids and phenolic acids on the inhibition of adipogenesis in 3T3-L1 adipocytes. J. Agric. Food Chem. 2007, 55, 8404–8410. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.Y.; Cosma, G.; Gardner, H.; Shi, X.; Castranova, V.; Vallyathan, V. Effect of antioxidant protection by p-coumaric acid on low-density lipoprotein cholesterol oxidation. American Journal of Physiology. Cell Physiol. 2000, 279, 954–960. [Google Scholar]
- Abdel-Wahab, M.H.; El-Mahdy, M.A.; Abd-Ellah, M.F.; Helal, G.K.; Khalifa, F.; Hamada, F.M. Influence of p-coumaric acid on doxorubicin-induced oxidative stress in rat’s heart. Pharmacol. Res. 2003, 48, 461–465. [Google Scholar] [CrossRef]
- Hudson, E.A.; Dinh, P.A.; Kokubun, T.; Simmonds, M.S.; Gescher, A. Characterization of potentially chemopreventive phenols in extracts of brown rice that inhibit the growth of human breast and colon cancer cells. Can. Epidemiol. Biomark. Prev. 2000, 9, 1163–1170. [Google Scholar]
- Lodovici, M.; Caldini, S.; Morbidelli, L.; Akpan, V.; Ziche, M.; Dolara, P. Protective effect of 4-coumaric acid from UVB ray damage in the rabbit eye. Toxicological 2009, 255, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Vauzour, D.; Corona, G.; Spencer, J.P. Caffeic acid, tyrosol and p-coumaric acid are potent inhibitors of 5-S-cysteinyldopamine induced neurotoxicity. Arch. Biochem. Biophys. 2010, 501, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Moon, J.H.; Seong, K.Y.; Park, K.H. Antimicrobial activity of 4-hydroxybenzoic acid and trans 4-hydroxycinnamic acid isolated and identified from rice hull. Biosci. Biotechnol. Biochem. 1998, 62, 2273–2276. [Google Scholar] [CrossRef] [PubMed]
- Luceri, C.; Guglielmi, F.; Lodovici, M.; Giannini, L.; Messerini, L.; Dolara, P. Plant phenolic 4-coumaric acid protects against intestinal inflammation in rats. Scand. J. Gastroenterol. 2004, 39, 1128–1133. [Google Scholar] [PubMed]
- Pragasam, S.J.; Murunikkara, V.; Sabina, E.P.; Rasool, M. Ameliorative effect of p-coumaric acid, a common dietary phenol, on adjuvant-induced arthritis in rats. Rheumatol. Int. 2013, 33, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Musaro, A.; Rosenthal, N. Regulation of muscle atrophy in aging and disease. Adv. Exp. Med. Biol. 2010, 694, 211–233. [Google Scholar] [PubMed]
- Buckingham, M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Megeney, L.A.; Rudnicki, M.A. Determination versus differentiation and the MyoD family of transcription factors. Biochem. Cell Biol. 1995, 73, 723–732. [Google Scholar] [CrossRef] [PubMed]
- De La Serna, I.L.; Carlson, K.A.; Imbalzano, A.N. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 2001, 2, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N.; Klein, W.H. Muscle minus MyoD. Dev. Biol. 1998, 202, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Kablar, B.; Krastel, K.; Tajbakhsh, S.; Rudnicki, M.A. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev. Biol. 2003, 258, 307–318. [Google Scholar] [CrossRef]
- Myer, A.; Olson, E.N.; Klein, W.H. MyoD cannot compensate for the absence of Myogenin during skeletal muscle differentiation in murine embryonic stem cells. Dev. Biol. 2001, 229, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Meadows, E.; Cho, J.H.; Flynn, J.M.; Klein, W.H. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev. Biol. 2008, 322, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Breitschopf, K.; Bengal, E.; Ziv, T.; Admon, A.; Ciechanover, A. A novel site for ubiquitination: The N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998, 17, 5964–5973. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Puri, P.L.; Hamamori, Y.; Ogryzko, V.; Chung, G.; Nakatani, Y.; Wang, J.Y.; Kedes, L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 1999, 4, 725–734. [Google Scholar] [CrossRef]
- Jo, C.; Cho, S.J.; Jo, S.A. Mitogen-activated protein kinase kinase 1 (MEK1) stabilizes MyoD through direct phosphorylation at tyrosine 156 during myogenic differentiation. J. Biol. Chem. 2011, 286, 18903–18913. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.M.; Bharathy, N.; Chung, T.K.; Kok, W.K.; Li, S.; Tan, Y.H.; Rao, V.K.; Gopinadhan, S.; Sartorelli, V.; Walsh, M.J.; et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. USA 2012, 109, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Kitzmann, M.; Carnac, G.; Vandromme, M.; Primig, M.; Lamb, N.J.; Fernandez, A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998, 142, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Blais, A.; Tsikitis, M.; Acosta-Alvear, D.; Sharan, R.; Kluger, Y.; Dynlacht, B.D. An initial blueprint for myogenic differentiation. Genes Dev. 2005, 19, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Solis, R.S.; Arias, E.B.; Cartee, G.D. Postcontraction insulin sensitivity: Relationship with contraction protocol, glycogen concentration, and 5 = AMP-activated protein kinase phosphorylation. J. Appl. Physiol. 2004, 96, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Ruderman, N.B.; Xu, X.J.; Nelson, L.; Cacicedo, J.M.; Saha, A.K.; Lan, F.; Ido, Y. AMPK and SIRT1: A long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 2010, 298, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Jeyabalan, J.; Shah, M.; Viollet, B.; Chenu, C. AMP-activated protein kinase pathway and bone metabolism. J. Endocrinol. 2012, 212, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Rattan, R.; Haq, E.; Khan, M.; Yasmin, R.; Won, J.S.; Key, L.; Singh, A.K.; Singh, I. AICAR inhibits adipocyte differentiation in 3T3L1 and restores metabolic alterations in diet-induced obesity mice model. Nutr. Metab. (Lond.) 2006, 3. [Google Scholar] [CrossRef]
- Pauly, M.; Daussin, F.; Burelle, Y.; Li, T.; Godin, R.; Fauconnier, J.; Koechlin-Ramonatxo, C.; Hugon, G.; Lacampagne, A.; Coisy-Quivy, M.; et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse dia-phragm. Am. J. Pathol. 2012, 181, 583–592. [Google Scholar]
- Fu, X.; Zhao, J.X.; Liang, J.; Zhu, M.J.; Viollet, B.; Du, M. AMP-activated protein kinase mediates Myogenin expression and myogenesis via histone deacetylase 5. Am. J. Physiol. Cell Physiol. 2013, 305, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhao, J.X.; Zhu, M.J.; Foretz, M.; Viollet, B.; Dodson, M.V.; Du, M. AMP-Activated Protein Kinase α1 but Not α 2 Catalytic Subunit Potentiates Myogenin Expression and Myogenesis. Mol. Cell Biol. 2013, 33, 4517–4525. [Google Scholar] [CrossRef] [PubMed]
- Couillard, C.; Mauriège, P.; Imbeault, P.; Prud’homme, D.; Nadeau, A.; Tremblay, A.; Bouchard, C.; Després, J.P. Hyperleptinemia is more closely associated with adipose cell hypertrophy than with adipose tissue hyperplasia. Int. J. Obes. Relat. Metab. Disord 2000, 24, 782–788. [Google Scholar] [CrossRef]
- Tontonoz, P.; Hu, E.; Graves, R.A.; Budavari, A.I.; Spiegelman, B.M. PPAR gamma 2: Tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994, 8, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Brun, R.P.; Kim, J.B.; Hu, E.; Altiok, S.; Spiegelman, B.M. Adipocyte differentiation: A transcriptional regulatory cascade. Curr. Opin. Cell Biol. 1996, 8, 826–832. [Google Scholar] [CrossRef]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar] [PubMed]
- Eberle, D.; Hegarty, B.; Bossard, P.; Ferre, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Spiegelman, B.M. A DD1/SREBP1 promote adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996, 10, 1096–1107. [Google Scholar]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Paulauskis, J.D.; Sul, H.S. Cloning and expression of mouse fatty acid synthase and other specific mRNAs Developmental and hormonal regulation in 3T3-L1 cells. J. Biol. Chem. 1988, 263, 7049–7054. [Google Scholar]
- Waite, M.; Wakil, S.J. Studies on the mechanism of action of acetyl coenzyme A carboxylase. I. Effect of isocitrate on the transcarboxylation step ofacetyl coenzyme A carboxylase. J. Biol. Chem. 1963, 238, 77–80. [Google Scholar]
- Naowaboot, J.; Chung, C.H.; Pannangpetch, P.; Choi, R.; Kim, B.H.; Lee, M.Y.; Kukongviriyapan, U. Mulberry leaf extract increases adiponectin in murine 3T3-L1 adipocytes. Nut. Res. 2012, 32, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.C.; Roh, S.G.; Hong, Y.H.; Shrestha, Y.B.; Hishikawa, D. The role of ghrelin and growth hormone secretagogues receptor on rat adipogenesis. Endocrinology 2003, 144, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the coumaric acid and cells lines (3T3-L1 and C2C12) are available from the authors.







© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilavenil, S.; Kim, D.H.; Srigopalram, S.; Arasu, M.V.; Lee, K.D.; Lee, J.C.; Lee, J.S.; Renganathan, S.; Choi, K.C. Potential Application of p-Coumaric Acid on Differentiation of C2C12 Skeletal Muscle and 3T3-L1 Preadipocytes—An in Vitro and in Silico Approach. Molecules 2016, 21, 997. https://doi.org/10.3390/molecules21080997
Ilavenil S, Kim DH, Srigopalram S, Arasu MV, Lee KD, Lee JC, Lee JS, Renganathan S, Choi KC. Potential Application of p-Coumaric Acid on Differentiation of C2C12 Skeletal Muscle and 3T3-L1 Preadipocytes—An in Vitro and in Silico Approach. Molecules. 2016; 21(8):997. https://doi.org/10.3390/molecules21080997
Chicago/Turabian StyleIlavenil, Soundharrajan, Da Hye Kim, Srisesharam Srigopalram, Mariadhas Valan Arasu, Kyung Dong Lee, Jeong Chae Lee, Jong Suk Lee, Senthil Renganathan, and Ki Choon Choi. 2016. "Potential Application of p-Coumaric Acid on Differentiation of C2C12 Skeletal Muscle and 3T3-L1 Preadipocytes—An in Vitro and in Silico Approach" Molecules 21, no. 8: 997. https://doi.org/10.3390/molecules21080997






