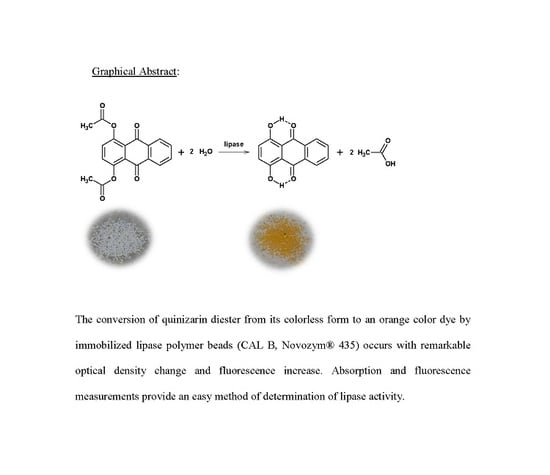
Monitoring the Activity of Immobilized Lipase with Quinizarin Diester Fluoro-Chromogenic Probe
Abstract
:1. Introduction
2. Experimental
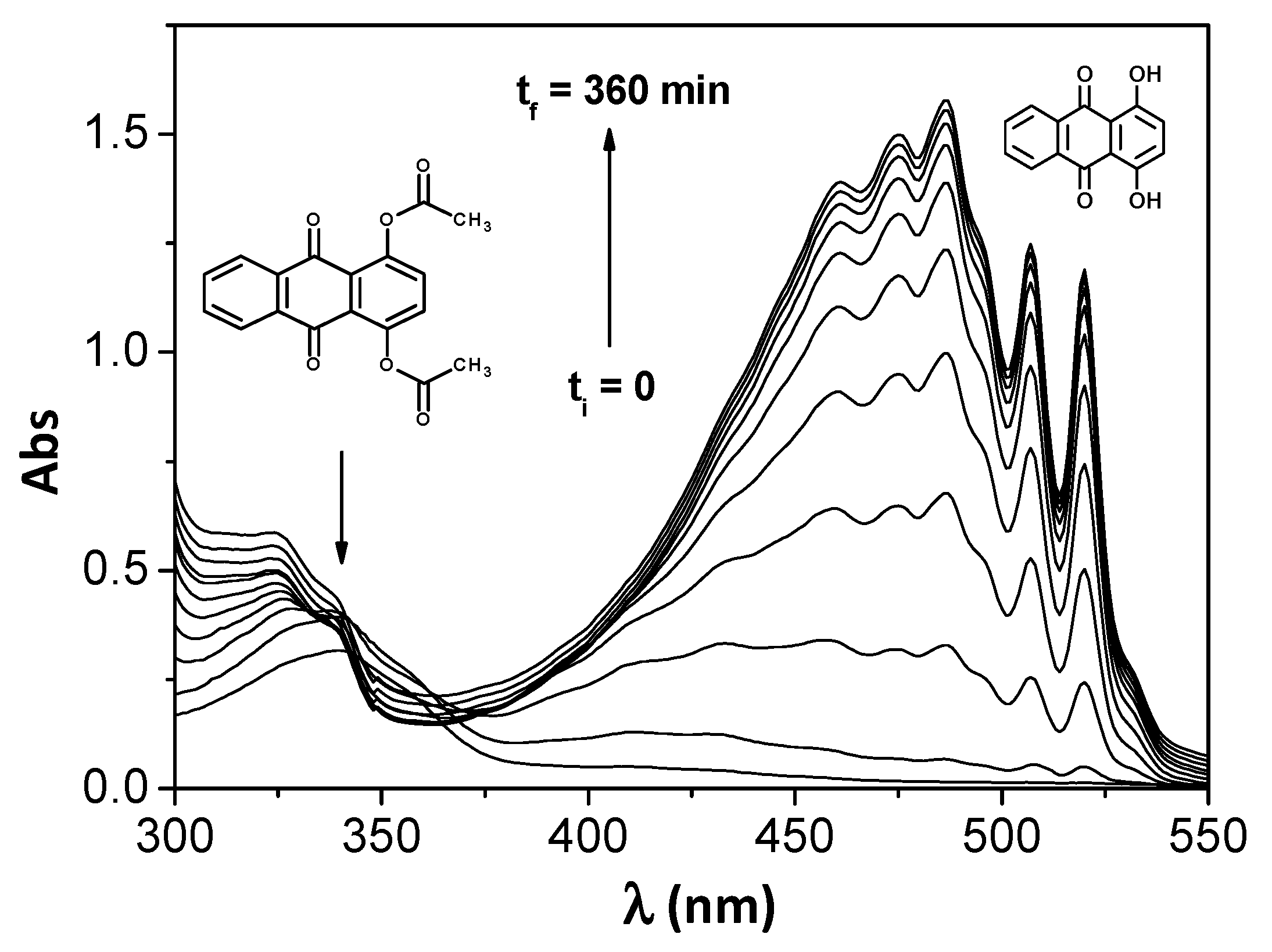
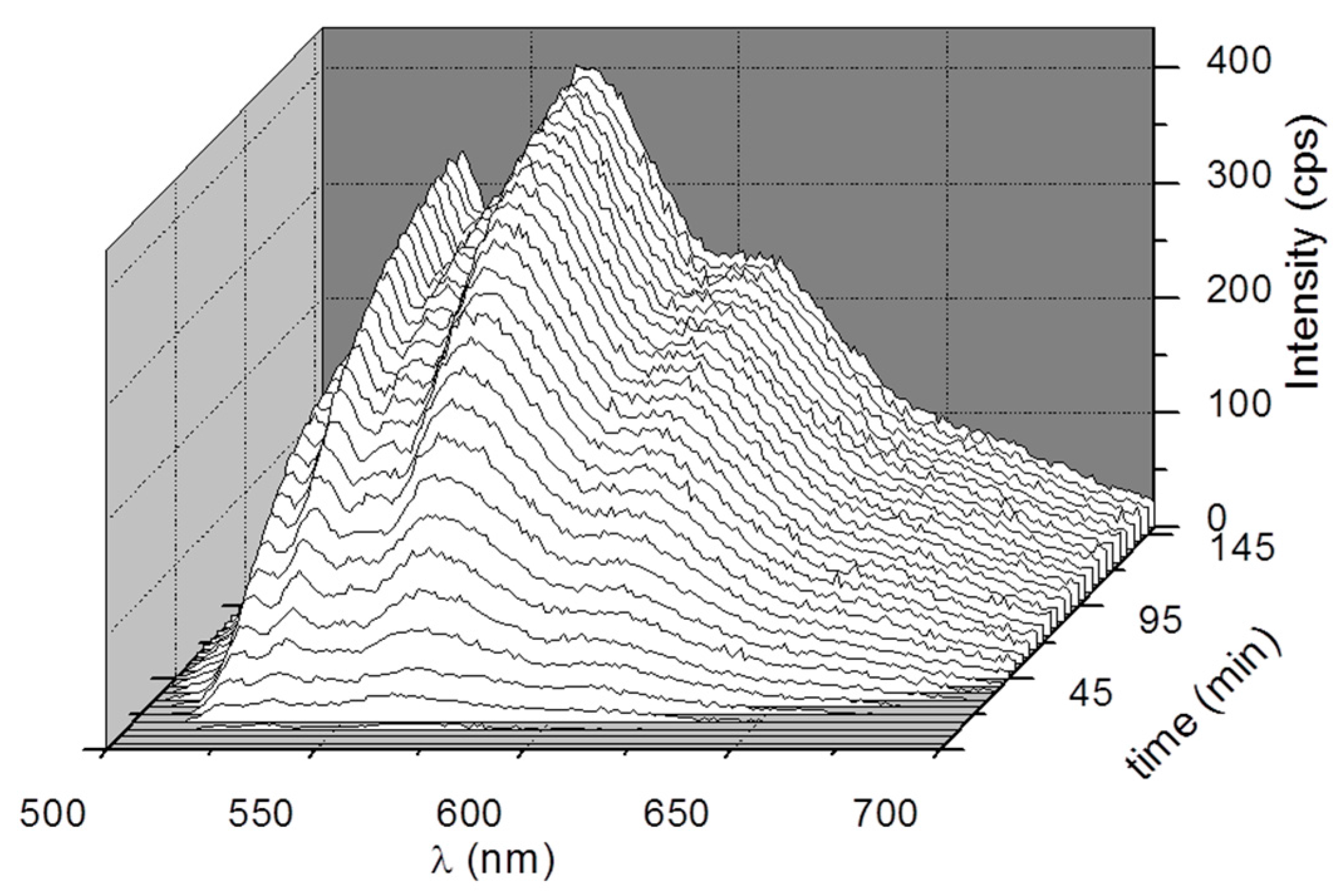
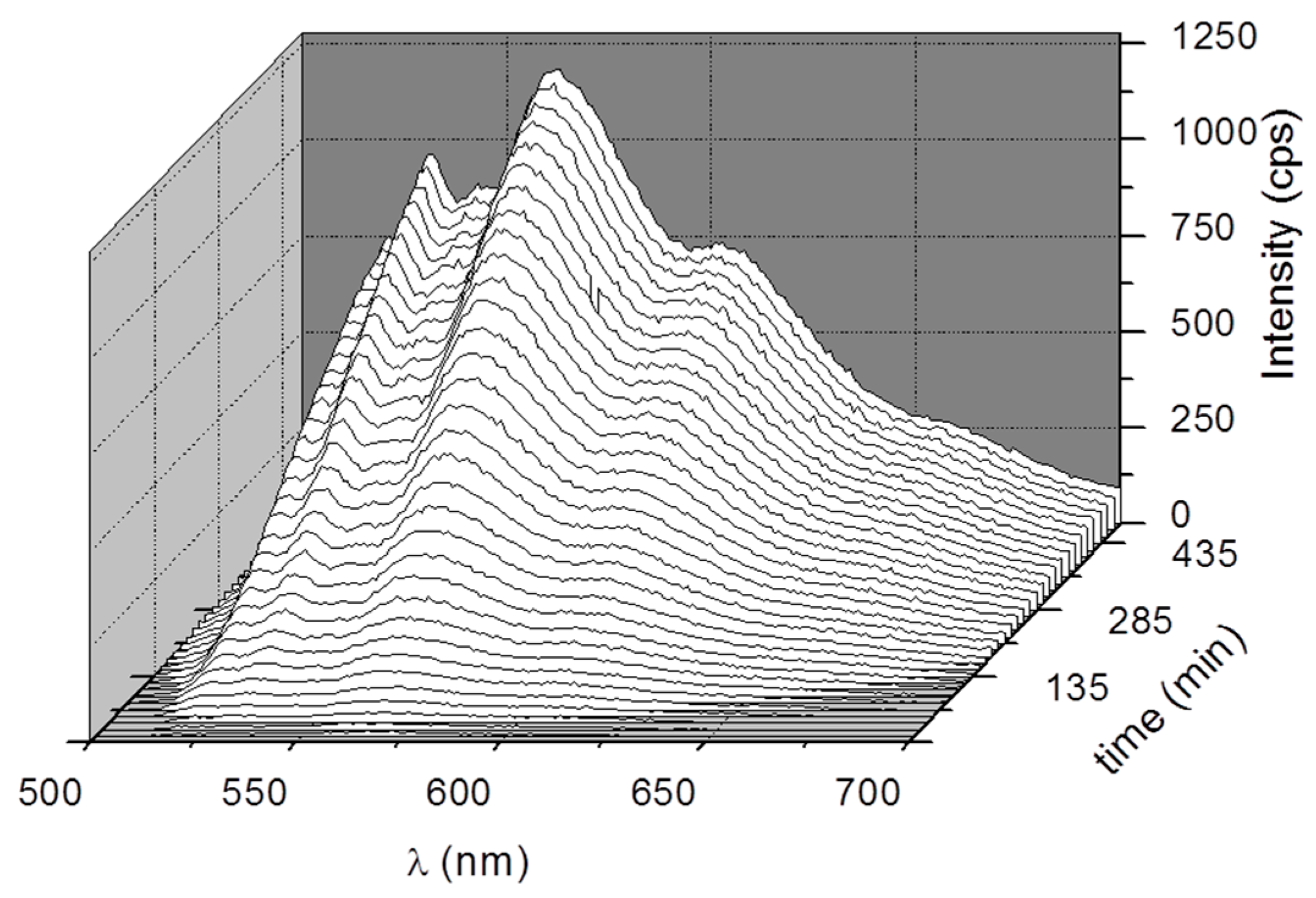
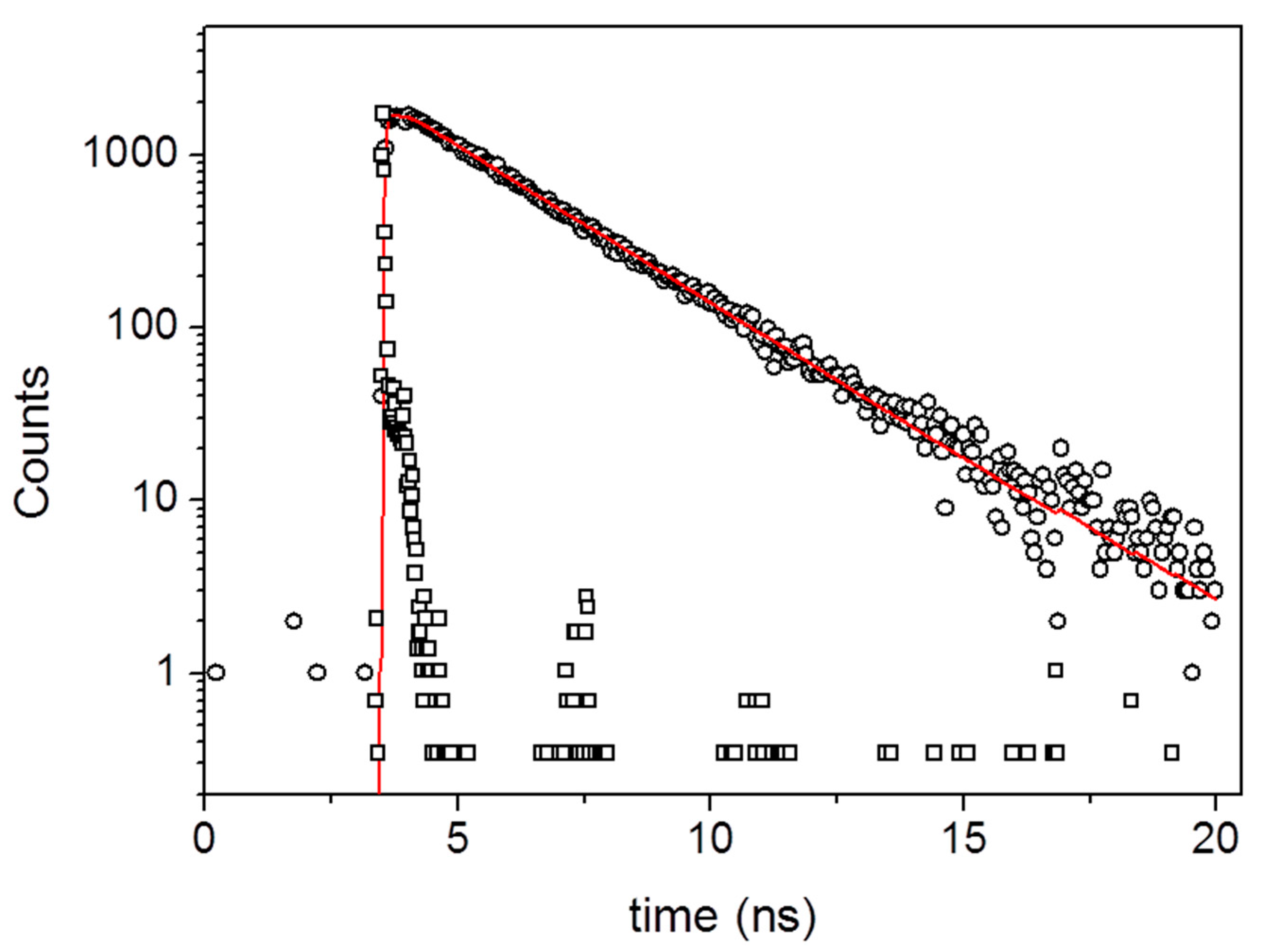
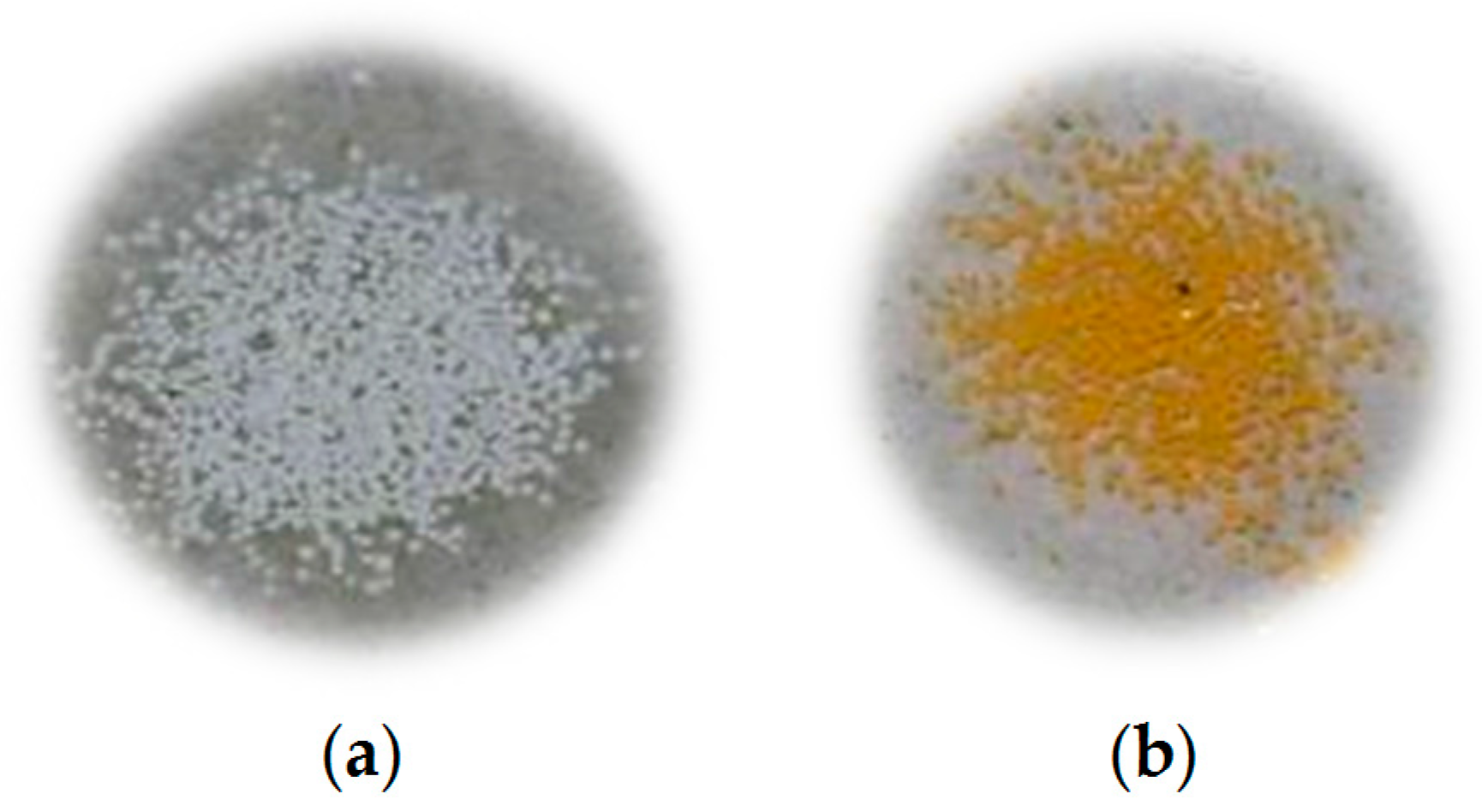
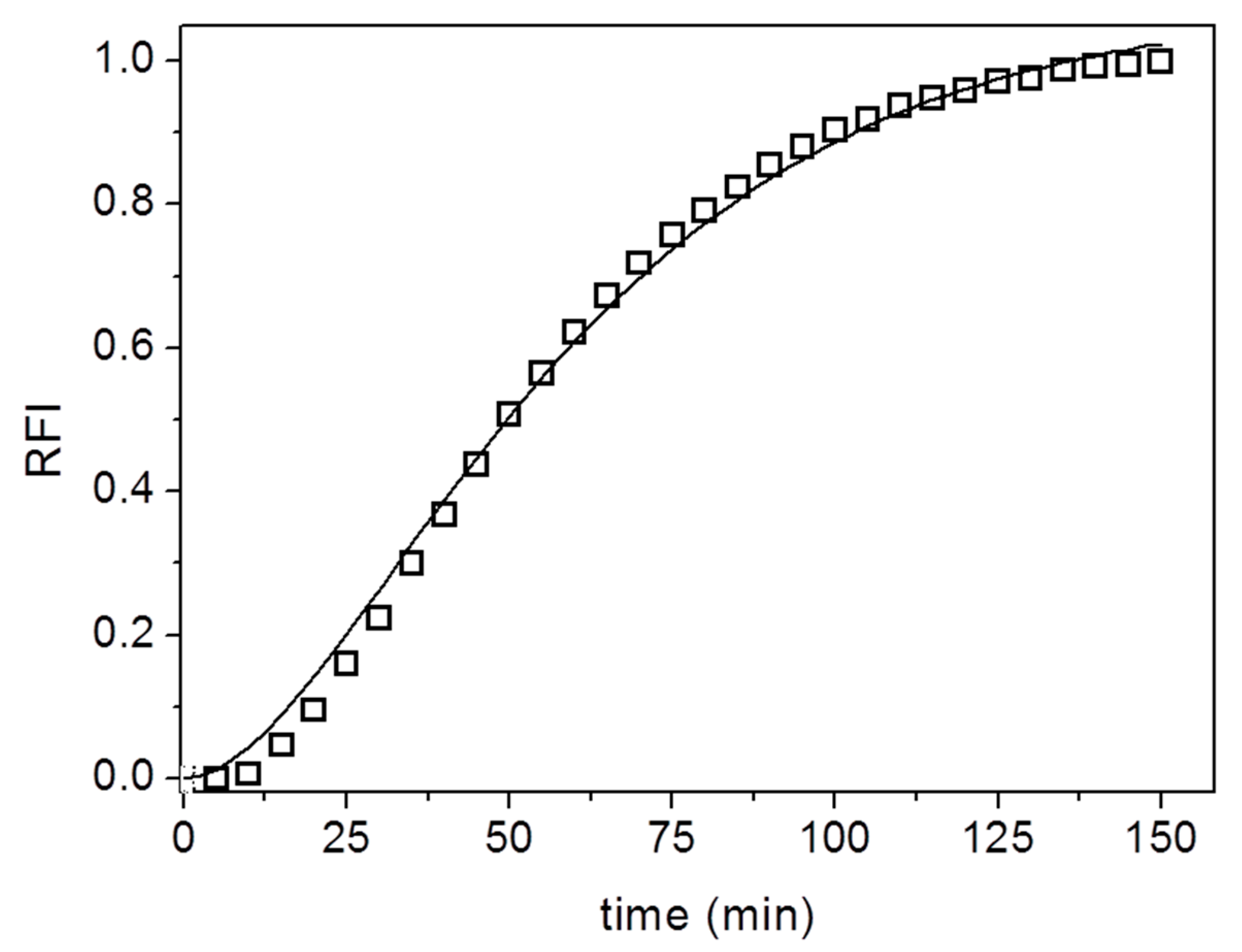
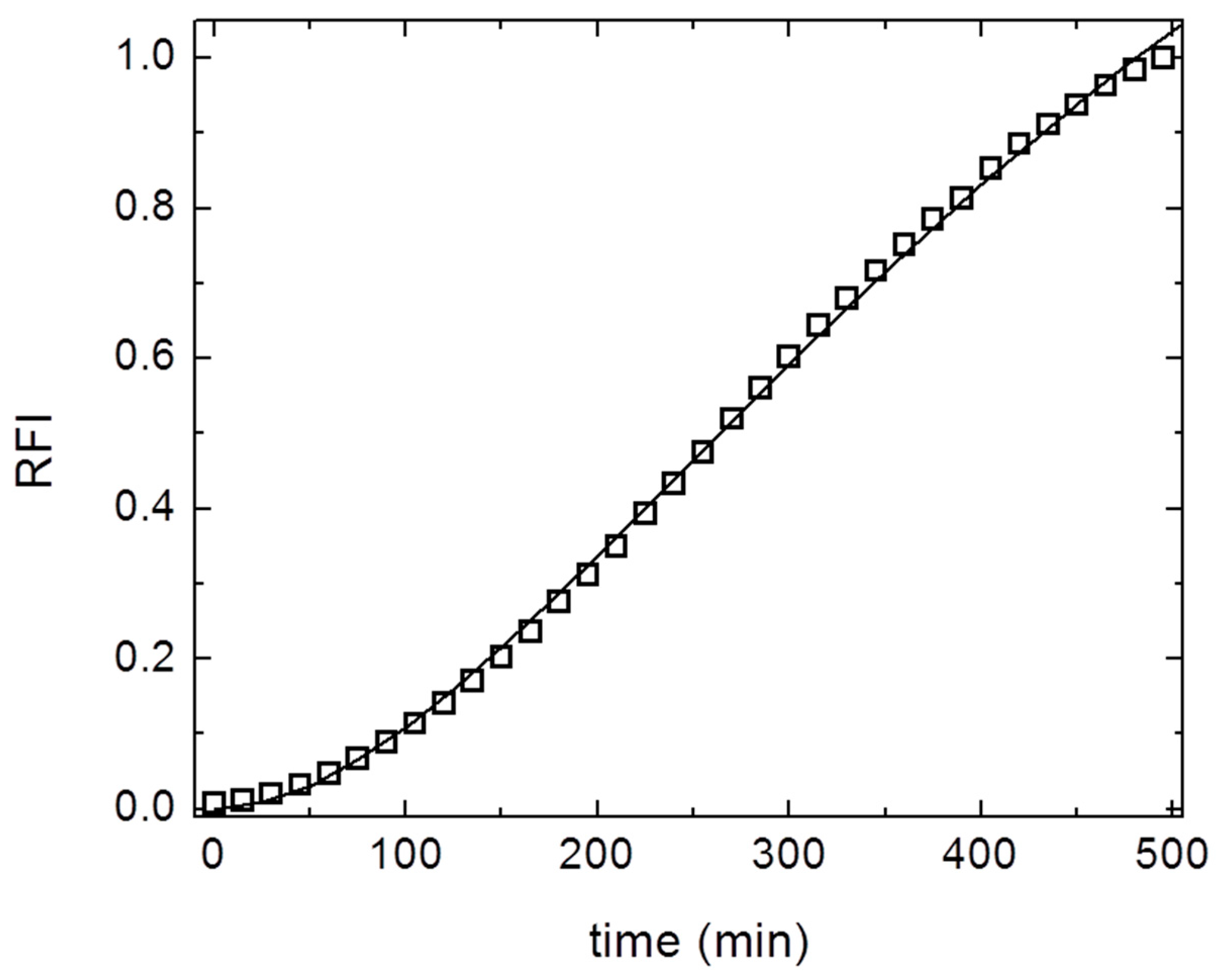
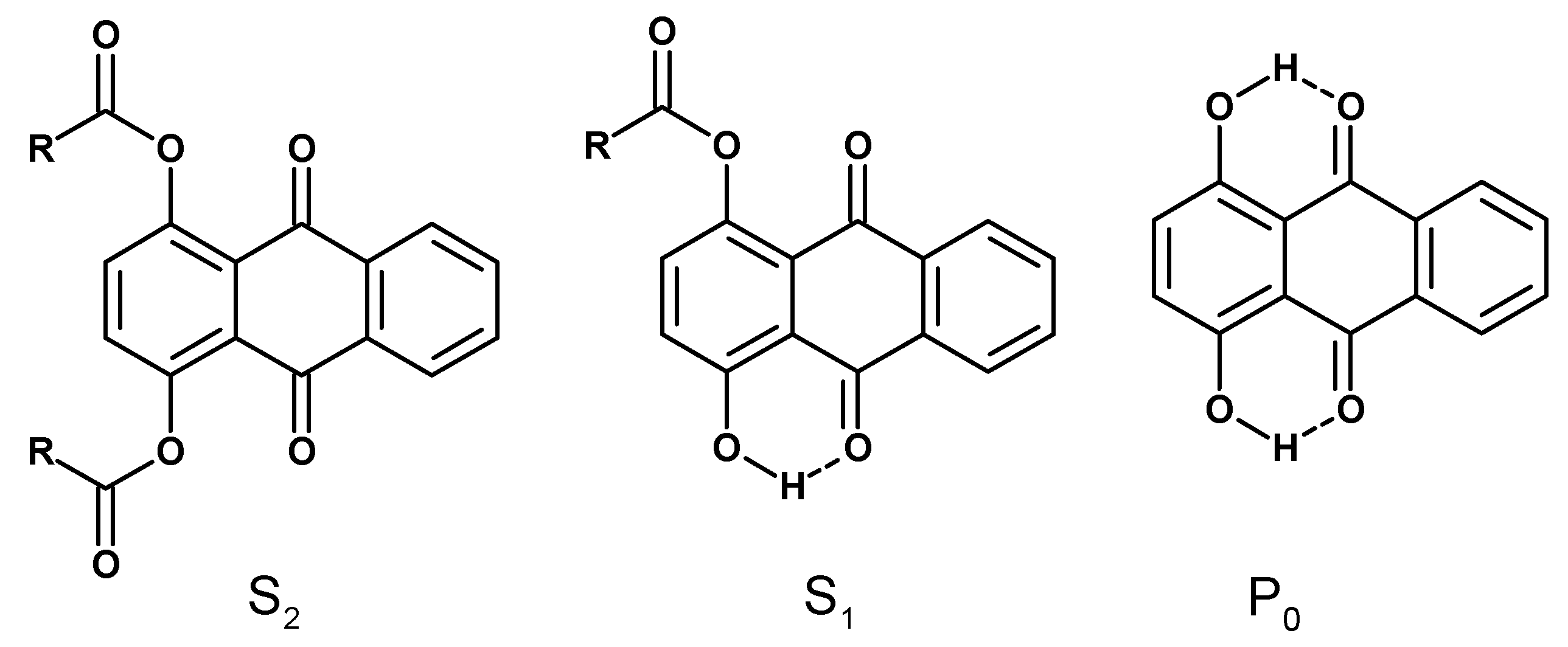
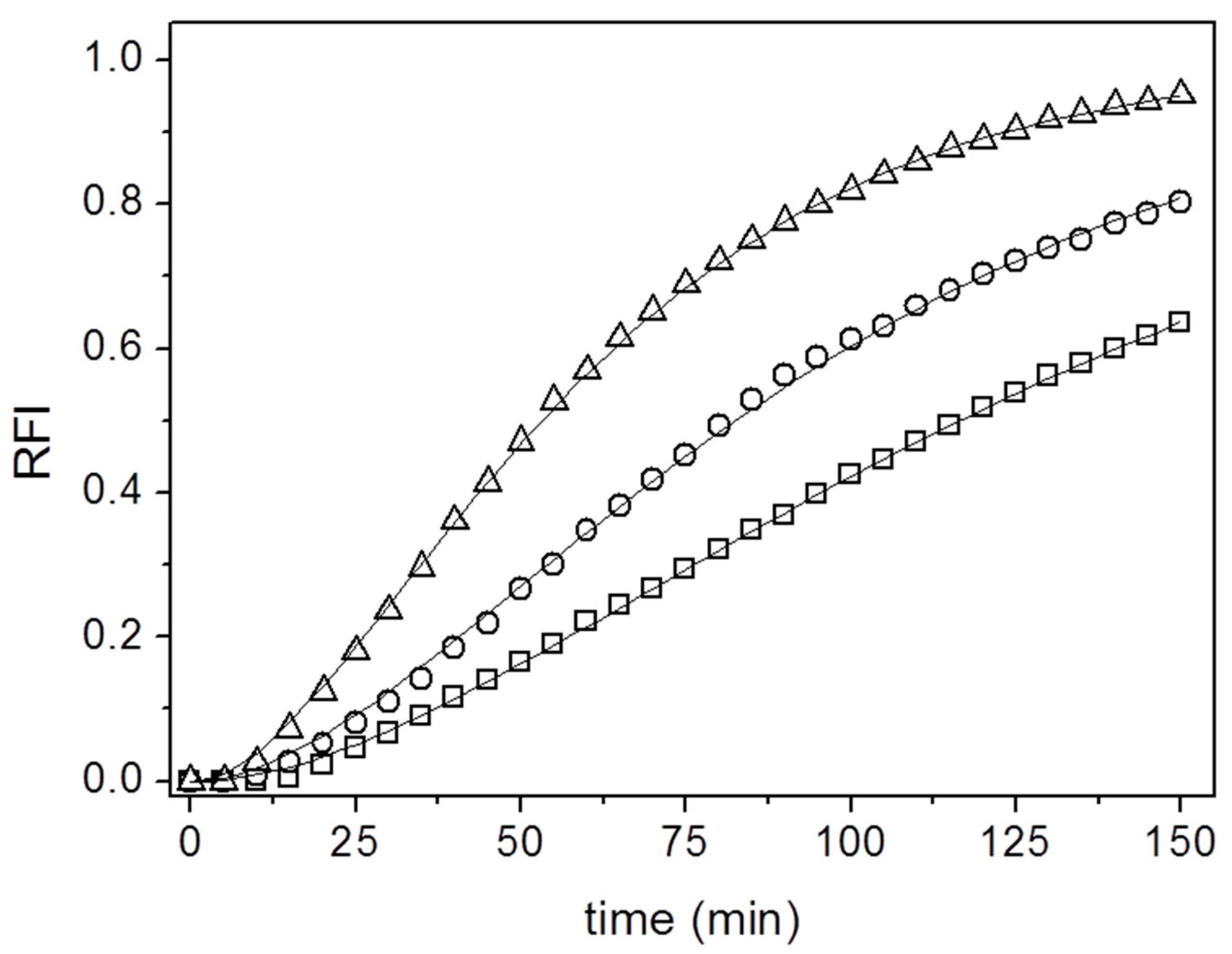
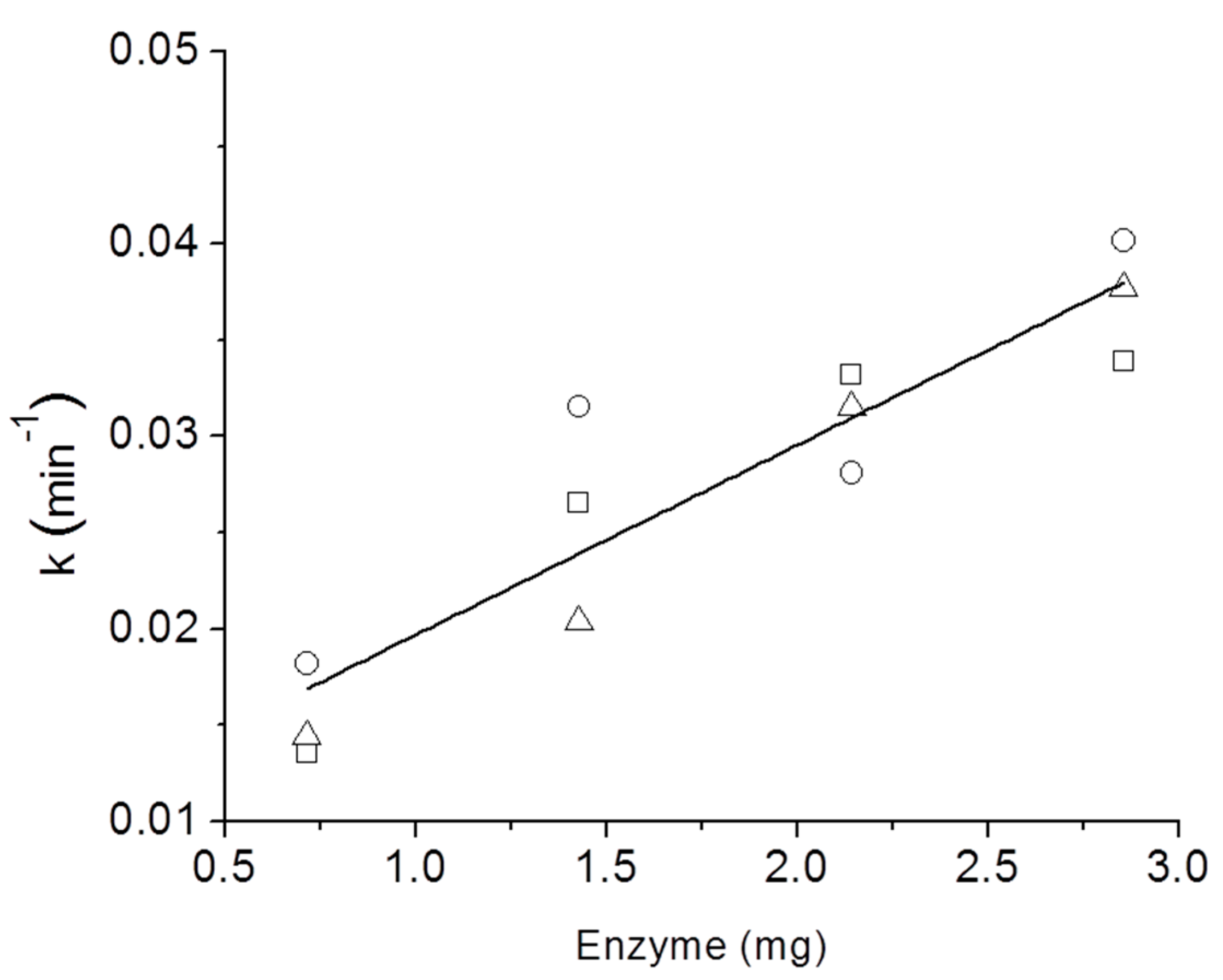
3. Results and Discussion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Villeneuve, P.; Muderhwa, J.M.; Graille, J.; Haas, M.J. Customizing lipases for biocatalysis: A survey of chemical, physical and molecular biological approaches. J. Mol. Catal. B Enzym. 2000, 9, 113–148. [Google Scholar] [CrossRef]
- Hoffmann, I.; Silva, V.D.; Nascimento, M.D. Enantioselective resolution of (R,S)-1-phenylethanol catalyzed by lipases immobilized in starch films. J. Braz. Chem. Soc. 2011, 22, 1559–1567. [Google Scholar] [CrossRef]
- Lima, G.V.; Silva, M.R.; Fonseca, T.S.; Lima, L.B.; Oliveira, M.C.F.; Lemos, T.L.G.; Zampieri, D.; Santos, J.C.S.; Rios, N.S.; Gonçalves, L.R.B.; et al. Chemoenzymatic synthesis of (S)-Pindolol using lipases. Appl. Catal. A 2017, 546, 7–14. [Google Scholar] [CrossRef]
- Khelassi, M.M.; Zaidi, A.; Zouioueche, L.A. CAL-B-Catalyzed deacylation of benzylic acetates: Effect of amines addition. Comparison of several approaches. Enzym. Microbial. Technol. 2017, 107, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Magadum, D.B.; Yadav, G.D. Enantioselective resolution of (R,S)-α-methyl-4-pyridinemethanolusing immobilized biocatalyst: Optimization and kinetic modeling. Biochem. Eng. J. 2017, 122, 152–158. [Google Scholar] [CrossRef]
- Al-Suhair, S. Production of biodiesel by lipase-catalyzed transesterification of vegetable oils: A kinetics study. Biotechnol. Prog. 2005, 21, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Gamba, M.; Lapis, A.A.M.; Dupont, J. Supported ionic liquid enzymatic catalysis for production of biodiesel. Adv. Synth. Catal. 2008, 350, 160–164. [Google Scholar] [CrossRef]
- Contesini, F.J.; Lopes, D.B.; Macedo, G.A.; Nascimento, M.D.; Carvalho, P.D. Aspergillus sp. Lipase: Potential biocatalyst for industrial use. J. Mol. Catal. B Enzym. 2010, 67, 163–171. [Google Scholar] [CrossRef]
- Verma, M.; Puri, M.; Barrow, C. Recent trends in nanomaterials immobilised enzymes for biofuel production. Crit. Rev. Biotechnol. 2016, 36, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Jin, Q.; Wang, X.; Wang, X. Preparation of medium and long chain triacylglycerols by lipase-catalyzed interesterification in a solvent-free system. Process Biochem. 2017, 54, 89–95. [Google Scholar] [CrossRef]
- Pascacio, V.G.T.; Ortíz, J.J.V.; Pérez, M.J.; Yates, M.; Sanchez, B.T.; Quintero, A.R.; Lafuente, R.F. Evaluation of different lipase biocatalysts in the production of biodiesel from used cooking oil: Critical role of the immobilization support. Fuel 2017, 200, 1–10. [Google Scholar] [CrossRef]
- Schöffer, J.N.; Matte, C.R.; Charqueiro, D.S.; Menezes, E.W.; Costa, T.M.H.; Benvenutti, E.V.; Rodrigues, R.C.; Hertz, P.F. Directed immobilization of CGTase: The effect of the enzyme orientation on the enzyme activity and its use in packed-bed reactor for continuous production of cyclodextrins. Process Biochem. 2017, 58, 120–127. [Google Scholar] [CrossRef]
- Su, F.; Li, G.L.; Fan, Y.L.; Yan, Y.J. Enhancing biodiesel production via a synergic effect between immobilized Rhizopus oryzae lipase and Novozym 435. Fuel Process. Technol. 2015, 137, 298–304. [Google Scholar] [CrossRef]
- Santos, J.C.S.; Rueda, N.; Gonçalves, L.R.B.; Lafuente, R.F. Tuning the catalytic properties of lipases immobilized on divinylsulfone activated agarose by altering its nanoenvironment. Enzym. Microb. Technol. 2015, 77, 1–77. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Canet, A.; Rivera, I.; Osório, N.M.; Sandoval, G.; Valero, F.; Dias, S.F. Biodiesel production from crude Jatropha oil catalyzed by non-commercial immobilized heterologous Rhizopus oryzae and Carica papaya lipases. Bioresour. Technol. 2016, 213, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Rios, N.S.; Pinheiro, M.P.; Santos, J.C.S.; Fonseca, T.S.; Lima, L.D.; Mattos, M.C.; Freire, D.M.G.; Júnior, I.J.S.; Aguado, E.R.; Gonçalves, L.R.B. Strategies of covalent immobilization of a recombinant Candidaantarctica lipase B on pore-expanded SBA-15 and its application in the kinetic resolution of (R,S)-Phenylethyl acetate. J. Mol. Catal. B Enzym. 2016, 133, 246–258. [Google Scholar] [CrossRef]
- Haugland, R.P.; Johnson, I.D. Detecting enzymes in living cells using fluorogenic substrates. J. Fluoresc. 1993, 3, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lauria, S.; Casati, S.; Ciuffreda, P. Synthesis and characterization of a new fluorogenic substrate for nonoacylglycerol lipase and application to inihibition studies. Anal. Bioanal. Chem. 2015, 407, 8163–8167. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, H.S. Fluorescence-based assays of lipases, phospholipases, and other lipolytic enzymes. Anal. Biochem. 1994, 219, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tallman, K.R.; Beatty, K.E. Far-red fluorogenic probes for esterase and lipase detection. ChemBioChem 2015, 16, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Prim, N.; Sánchez, M.; Ruiz, C.; Javier Pastor, F.I.; Diaz, P. Use of methylumbeliferyl-derivative substrates for lipase activity characterization. J. Mol. Catal. B Enzym. 2003, 22, 339–346. [Google Scholar] [CrossRef]
- Flomenbom, O.; Velonia, K.; Loss, D.; Masuo, S.; Cotlet, M.; Engelborghs, Y.; Hofkens, J.; Rowan, A.E.; Nolte, R.J.M.; van der Auweraer, M.; et al. Stretched exponential decay and correlations in the catalystic activity of fluctuating single lipase molecules. Proc. Nat. Acad. Sci. USA 2005, 102, 2368–2372. [Google Scholar] [CrossRef] [PubMed]
- Liebherr, R.B.; Gorris, H.H. Enzyme molecules in solitary confinement. Molecules 2014, 19, 14417–14445. [Google Scholar] [CrossRef] [PubMed]
- Ursoiu, A.; Paul, C.; Kurtán, T.; Péter, F. Sol-gel entrapped candida antarctica lipase B—A biocatalyst with excelente stability for kinetic resolution of secondary alcohols. Molecules 2012, 17, 13045–13061. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Babiak, P.; Reymond, J.-L. New nonofunctionalized fluorescein derivatives for the efficient high-throughput screening of lipases and esterases in aqueous media. Helv. Chim. Acta 2006, 89, 404–415. [Google Scholar] [CrossRef]
- Mantovani, S.M.; de Oliveira, L.G.; Marsaioli, A.J. Whole cell quick E for epoxide hydrolases screening using fluorescente probes. J. Mol. Catal. B Enzym. 2008, 52, 173–177. [Google Scholar] [CrossRef]
- Mantovani, S.M.; de Oliveira, L.G.; Marsaioli, A.J. Esterase screening using whole cells of brazilian soil microorganisms. J. Braz. Chem. Soc. 2010, 21, 1484–1489. [Google Scholar] [CrossRef]
- Sabatini, C.A.; Gehlen, M.H. Enzymatic hydrolysis of quinizarin diester by lipase in silica nanoparticles investigated by fluorescence microscopy. J. Nanopart. Res. 2014, 16, 2093. [Google Scholar] [CrossRef]
- Pereira, R.V.; Gehlen, M.H. Photoinduced intramolecular charge transfer in 9-aminoacridinium derivatives assisted by intramolecular H-bond. J. Phys. Chem. A 2006, 110, 7539–7546. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Kang, J.H.; Han, D.-K.; Lee, C.W.; Ahn, K.D. A novel precursor for color and fluorescence imaging. Chem. Mater. 1998, 10, 2332–2334. [Google Scholar] [CrossRef]
- Ahn, K.D.; Yoo, K.W.; Soh, J.H.; Kang, J.H. Fluorescent photoimaging with polymer having protected quinizarin dye precursors by a dry process based on chemical amplification. React. Funct. Polym. 2009, 69, 111–116. [Google Scholar] [CrossRef]
- Pal, H.; Palit, D.K.; Mukherjee, T.; Mittal, J.P. Interaction of the excited singlet state of disubstituted anthraquinones with aromatic hydrocarbons: A fluorescence-quenching study. Chem. Phys. Lett. 1990, 173, 354–359. [Google Scholar] [CrossRef]
- Palit, D.K.; Pal, H.; Mukherjee, T.; Mittal, J.P. Photodynamics of the S1 state of some hydroxyl-and amino-substituted naphthoquinones and anthraquinones. J. Chem. Soc. Faraday Trans. 1990, 86, 3861–3869. [Google Scholar] [CrossRef]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from candida Antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Costa, L.; Brissos, V.; Lemos, F.; Ribeiro, F.R.; Cabral, J.M.S. Comparing the effect of immobilization methods on the activity of lipase biocatalysts in ester hydrolysis. Bioprocess Biosyst. Eng. 2008, 31, 323–327. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
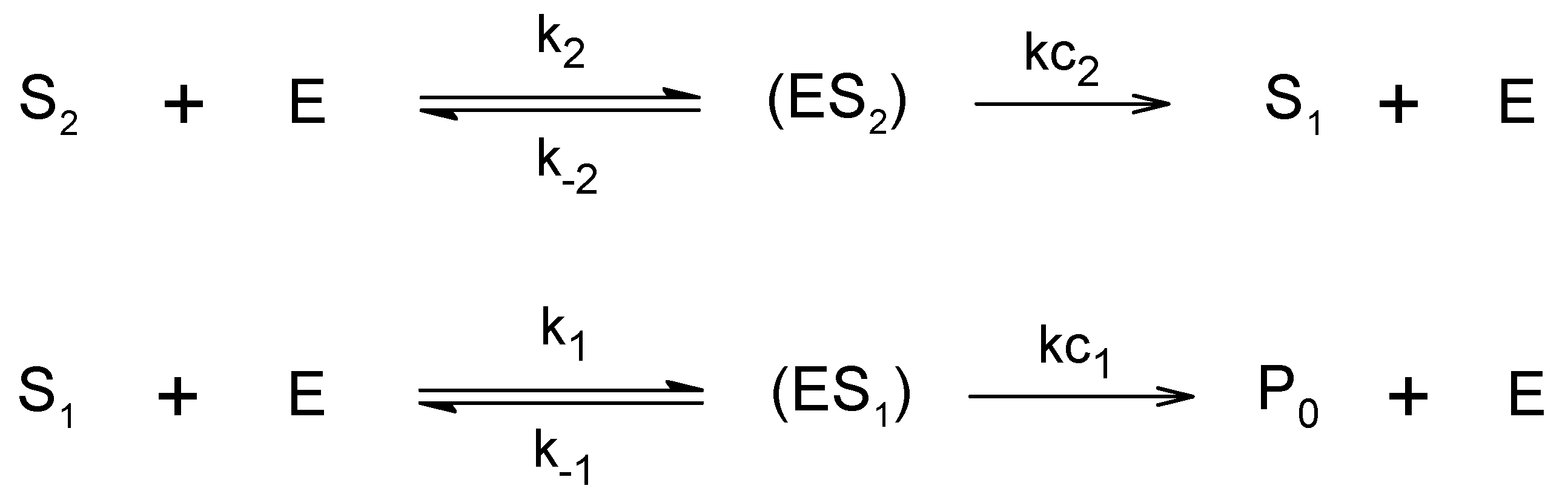
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aparecida Sabatini, C.; Massucatto dos Santos, D.; Matos de Oliveira da Silva, S.; Henrique Gehlen, M. Monitoring the Activity of Immobilized Lipase with Quinizarin Diester Fluoro-Chromogenic Probe. Molecules 2017, 22, 2136. https://doi.org/10.3390/molecules22122136
Aparecida Sabatini C, Massucatto dos Santos D, Matos de Oliveira da Silva S, Henrique Gehlen M. Monitoring the Activity of Immobilized Lipase with Quinizarin Diester Fluoro-Chromogenic Probe. Molecules. 2017; 22(12):2136. https://doi.org/10.3390/molecules22122136
Chicago/Turabian StyleAparecida Sabatini, Carolina, Denis Massucatto dos Santos, Sabrina Matos de Oliveira da Silva, and Marcelo Henrique Gehlen. 2017. "Monitoring the Activity of Immobilized Lipase with Quinizarin Diester Fluoro-Chromogenic Probe" Molecules 22, no. 12: 2136. https://doi.org/10.3390/molecules22122136