Domain IV of Annexin A5 Is Critical for Binding Calcium and Guarantees Its Maximum Binding to the Phosphatidylserine Membrane
Abstract
:1. Introduction
2. Results and Discussion
2.1. Successful Purification of AnxA5-Related Proteins
2.2. Truncation of Domain IV of AnxA5 Destroys Its Calcium-Binding Ability and Impairs Its Affinity for PS Liposome
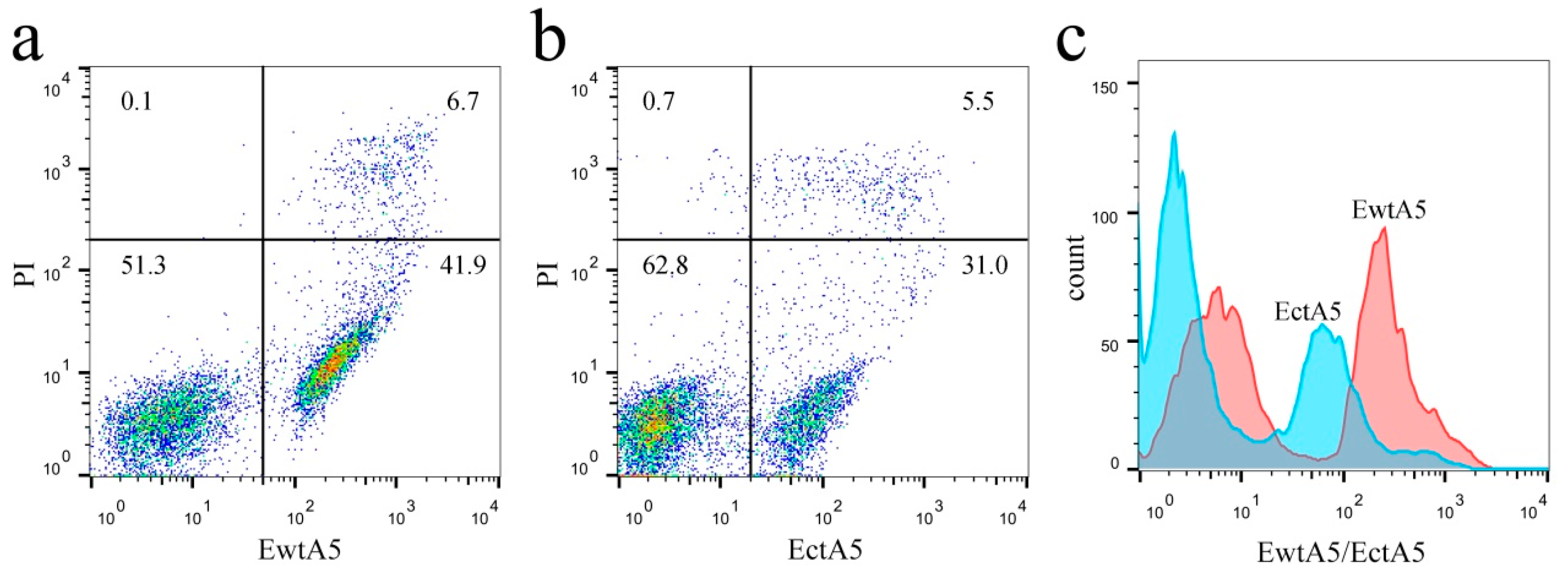
2.3. Truncation of Domain IV of AnxA5 Impairs Its Ability to Label Apoptotic Cells
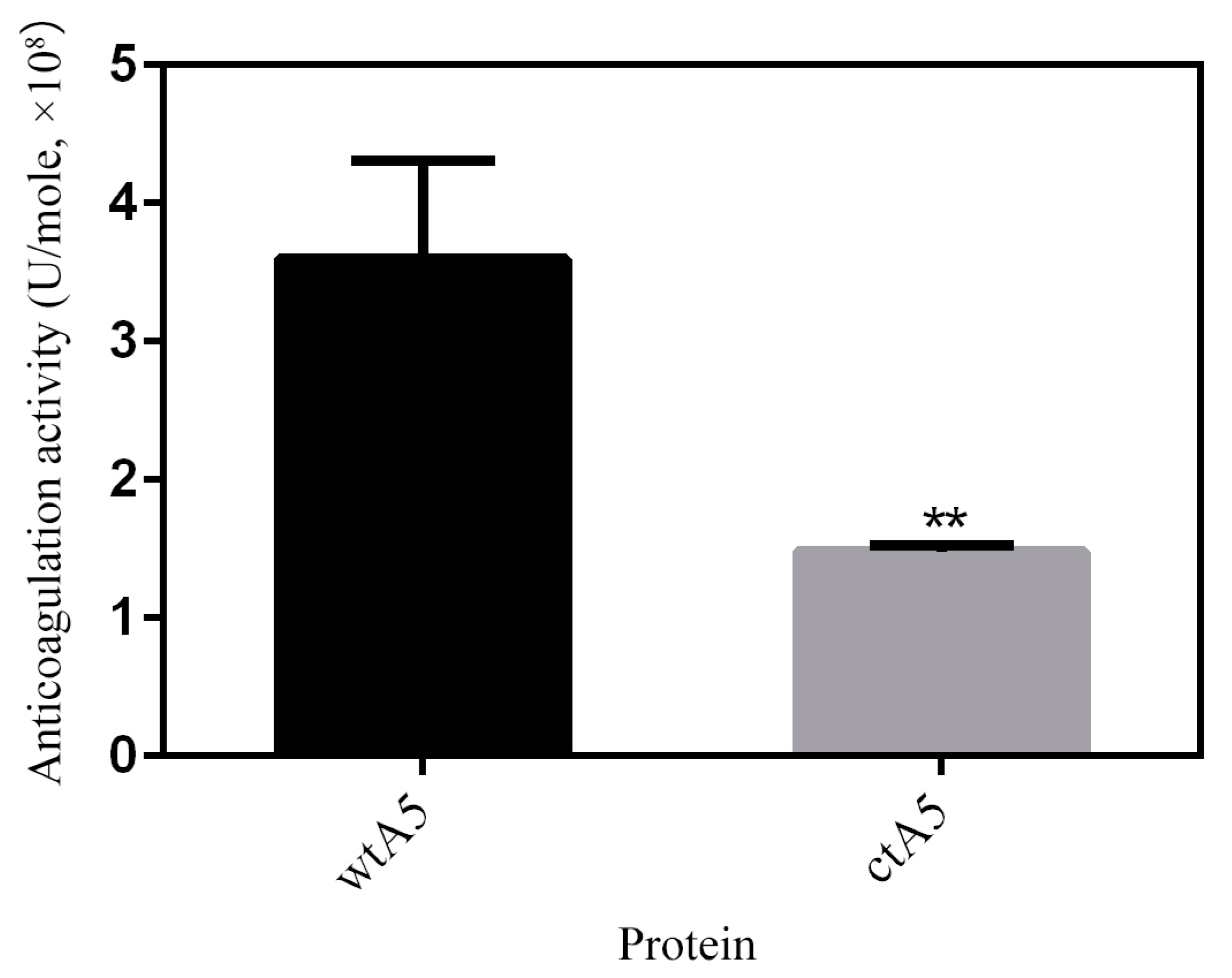
2.4. Truncation of Domain IV Impairs the Anticoagulation Activity of AnxA5
3. Materials and Methods
3.1. Construction of EGFP-AnxA5 Fusion Protein Expression Vectors
3.2. Construction of Intein-AnxA5 Fusion Protein Expression Vectors
3.3. Expression and Purification of AnxA5 and Related Proteins
3.4. SDS-PAGE Analysis
3.5. Isothermal Titration Calorimetry Analysis
3.6. Apoptosis Detection by Flow Cytometry
3.7. Anticoagulation Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lizarbe, M.A.; Barrasa, J.I.; Olmo, N.; Gavilanes, F.; Turnay, J. Annexin-phospholipid interactions. Functional implications. Int. J. Mol. Sci. 2013, 14, 2652–2683. [Google Scholar] [CrossRef] [PubMed]
- Garnier, B.; Bouter, A.; Gounou, C.; Petry, K.G.; Brisson, A.R. Annexin A5-Functionalized Liposomes for Targeting Phosphatidylserine-Exposing Membranes. Bioconjug. Chem. 2009, 20, 2114–2122. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.; Rentero, C.; Cairns, R.; Tebar, F.; Enrich, C.; Grewal, T. Annexins—Scaffolds modulating PKC localization and signaling. Cell Signal. Technol. 2014, 26, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Turnay, J.; Olmo, N.; Gasset, M.; Iloro, I.; Arrondo, J.L.; Lizarbe, M.A. Calcium-dependent conformational rearrangements and protein stability in chicken annexin A5. Biophys. J. 2002, 83, 2280–2291. [Google Scholar] [CrossRef]
- Koopman, G.; Reutelingsperger, C.P.; Kuijten, G.A.; Keehnen, R.M.; Pals, S.T.; van Oers, M.H. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 1994, 84, 1415–1420. [Google Scholar] [PubMed]
- Boersma, H.H.; Kietselaer, B.; Stolk, L.M.L.; Bennaghmouch, A.; Hofstra, L.; Narula, J.; Heidendal, G.A.K.; Reutelingsperger, C.P.M. Past, present, and future of annexin A5: From protein discovery to clinical applications. J. Nucl. Med. 2005, 46, 2035–2050. [Google Scholar] [PubMed]
- Demchenko, A.P. Beyond annexin V: Fluorescence response of cellular membranes to apoptosis. Cytotechnology 2013, 65, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Bouter, A.; Carmeille, R.; Gounou, C.; Bouvet, F.; Degrelle, S.A.; Evain-Brion, D.; Brisson, A.R. Review: Annexin-A5 and cell membrane repair. Placenta 2015, 36, S43–S49. [Google Scholar] [CrossRef] [PubMed]
- Thiagarajan, P.; Benedict, C.R. Inhibition of arterial thrombosis by recombinant annexin V in a rabbit carotid artery injury model. Circulation 1997, 96, 2339–2347. [Google Scholar] [CrossRef] [PubMed]
- Ewing, M.M.; de Vries, M.; Nordzell, M.; Jukema, J.W.; Frostegard, J.; Pettersson, K.; Quax, P. Annexin A5 Reduces Inflammation Mediated Vascular Remodelling and Post-interventional Atherosclerosis and Improves Vascular Function in Hypercholesterolemic Mice. Circulation 2009, 120, S1113. [Google Scholar]
- Davis, B.M.; Normando, E.M.; Guo, L.; Turner, L.A.; Nizari, S.; O’Shea, P.; Moss, S.E.; Somavarapu, S.; Cordeiro, M.F. Topical delivery of Avastin to the posterior segment of the eye in vivo using annexin A5-associated liposomes. Small 2014, 10, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Van Rite, B.D.; Harrison, R.G. Annexin V-targeted enzyme prodrug therapy using cytosine deaminase in combination with 5-fluorocytosine. Cancer Lett. 2011, 307, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Hu, M.; Tang, B.; Liu, X.; Zhuang, H.; Yang, J.; Hua, Z.C. Annexin V-TRAIL fusion protein is a more sensitive and potent apoptotic inducer for cancer therapy. Sci. Rep. 2013, 3, 3565. [Google Scholar] [CrossRef] [PubMed]
- Belhocine, T.Z.; Blankenberg, F.G.; Kartachova, M.S.; Stitt, L.W.; Vanderheyden, J.-L.; Hoebers, F.J.P.; Van de Wiele, C. Tc-99m-Annexin A5 quantification of apoptotic tumor response: A systematic review and meta-analysis of clinical imaging trials. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 2083–2097. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kondo, S.; Mizuno, K.; Yano, W.; Nakao, H.; Hattori, Y.; Kimura, K.; Nishida, T. Promotion of corneal epithelial wound healing in vitro and in vivo by annexin A5. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1862–1868. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Van Genderen, H.O.; Kenis, H.; Hofstra, L.; Narula, J.; Reutelingsperger, C.P. Extracellular annexin A5: Functions of phosphatidylserine-binding and two-dimensional crystallization. Biochim. Biophys. Acta 2008, 1783, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Burgmaier, M.; Schutters, K.; Willems, B.; van der Vorst, E.P.; Kusters, D.; Chatrou, M.; Norling, L.; Biessen, E.A.; Cleutjens, J.; Perretti, M.; et al. AnxA5 reduces plaque inflammation of advanced atherosclerotic lesions in apoE(−/−) mice. J. Cell. Mol. Med. 2014, 18, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Montaville, P.; Neumann, J.M.; Russo-Marie, F.; Ochsenbein, F.; Sanson, A. A new consensus sequence for phosphatidylserine recognition by annexins. J. Biol. Chem. 2002, 277, 24684–24693. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mao, Y.; Yang, J.; Zhang, T.; Zhao, L.; Yu, K.; Zheng, M.; Jiang, H.; Yang, H. Characterizing the binding of annexin V to a lipid bilayer using molecular dynamics simulations. Proteins 2014, 82, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Mira, J.P.; Dubois, T.; Oudinet, J.P.; Lukowski, S.; Russo-Marie, F.; Geny, B. Inhibition of cytosolic phospholipase A2 by annexin V in differentiated permeabilized HL-60 cells. Evidence of crucial importance of domain I type II Ca2+-binding site in the mechanism of inhibition. J. Biol. Chem. 1997, 272, 10474–10482. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Smith, C.; Hsieh, H.Y.; Gibson, D.F.; Tait, J.F. Essential role of B-helix calcium binding sites in annexin V-membrane binding. J. Biol. Chem. 2004, 279, 40351–40357. [Google Scholar] [CrossRef] [PubMed]
- Somarajan, S.R.; Al-Asadi, F.; Ramasamy, K.; Pandranki, L.; Baseman, J.B.; Kannan, T.R. Annexin A2 mediates Mycoplasma pneumoniae community-acquired respiratory distress syndrome toxin binding to eukaryotic cells. mBio 2014, 5, e01497-14. [Google Scholar] [CrossRef] [PubMed]
- Sable, C.L.; Riches, D.W. Cloning and functional activity of a novel truncated form of annexin IV in mouse macrophages. Biochem. Biophys. Res. Commun. 1999, 258, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Sohma, H.; Creutz, C.E.; Saitoh, M.; Sano, H.; Kuroki, Y.; Voelker, D.R.; Akino, T. Characterization of the Ca2+-dependent binding of annexin IV to surfactant protein A. Biochem. J. 1999, 341, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Pons, M.; Grewal, T.; Rius, E.; Schnitgerhans, T.; Jackle, S.; Enrich, C. Evidence for the Involvement of annexin 6 in the trafficking between the endocytic compartment and lysosomes. Exp. Cell Res. 2001, 269, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Ying, Y.; Anderson, R.G. Annexin VI-mediated loss of spectrin during coated pit budding is coupled to delivery of LDL to lysosomes. J. Cell Biol. 1998, 142, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.C.; Chen, D.C.P. Novel Short Isoform of Annexin A10 at Chromosome 4q, Termed Annexin 10s (ANXA10s) and Methods of Use. U.S. Patent 7,495,073, 24 February 2009. [Google Scholar]
- Swaggart, K.A.; Demonbreun, A.R.; Vo, A.H.; Swanson, K.E.; Kim, E.Y.; Fahrenbach, J.P.; Holley-Cuthrell, J.; Eskin, A.; Chen, Z.; Squire, K.; et al. Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair. Proc. Natl. Acad. Sci. USA 2014, 111, 6004–6009. [Google Scholar] [CrossRef] [PubMed]
- Marder, L.S.; Lunardi, J.; Renard, G.; Rostirolla, D.C.; Petersen, G.O.; Nunes, J.E.; de Souza, A.P.; de O Dias, A.C.; Chies, J.M.; Basso, L.A.; et al. Production of recombinant human annexin V by fed-batch cultivation. BMC Biotechnol. 2014, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Coxon, K.M.; Duggan, J.; Cordeiro, M.F.; Moss, S.E. Purification of annexin V and its use in the detection of apoptotic cells. Methods Mol. Biol. 2011, 731, 293–308. [Google Scholar] [PubMed]
- Rand, J.H.; Wu, X.X.; Quinn, A.S.; Taatjes, D.J. Resistance to annexin A5 anticoagulant activity: A thrombogenic mechanism for the antiphospholipid syndrome. Lupus 2008, 17, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Cornely, R.; Rentero, C.; Enrich, C.; Grewal, T.; Gaus, K. Annexin A6 is an organizer of membrane microdomains to regulate receptor localization and signalling. IUBMB Life 2011, 63, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.Y.; Ou, Y.H.; Chen, W.J.; Li, H.Y.; Liu, S.H.; Pan, H.W.; Lai, P.L.; Jeng, Y.M.; Chen, D.C.; Hsu, H.C. Aberrant expressions of annexin A10 short isoform, osteopontin and alpha-fetoprotein at chromosome 4q cooperatively contribute to progression and poor prognosis of hepatocellular carcinoma. Int. J. Oncol. 2005, 26, 1053–1061. [Google Scholar] [PubMed]
- Morgan, R.O.; Martin-Almedina, S.; Iglesias, J.M.; Gonzalez-Florez, M.I.; Fernandez, M.P. Evolutionary perspective on annexin calcium-binding domains. Biochim. Biophys. Acta 2004, 1742, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Aukrust, I.; Evensen, L.; Hollas, H.; Berven, F.; Atkinson, R.A.; Trave, G.; Flatmark, T.; Vedeler, A. Engineering, biophysical characterisation and binding properties of a soluble mutant form of annexin A2 domain IV that adopts a partially folded conformation. J. Mol. Biol. 2006, 363, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Raddum, A.M.; Evensen, L.; Hollas, H.; Grindheim, A.K.; Lorens, J.B.; Vedeler, A. Domains I and IV of annexin A2 affect the formation and integrity of in vitro capillary-like networks. PLoS ONE 2013, 8, e60281. [Google Scholar] [CrossRef]
- Aukrust, I.; Hollas, H.; Strand, E.; Evensen, L.; Trave, G.; Flatmark, T.; Vedeler, A. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J. Mol. Biol. 2007, 368, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, L.; Chen, D.; Pi, Y.; Zhou, W.; Xiong, X.; Ren, Y.; Lai, Y.; Hua, Z. Quantitative analysis of annexin V-membrane interaction by flow cytometry. Eur. Biophys. J. Biophys. Lett. 2015, 44, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Gauer, J.W.; Knutson, K.J.; Jaworski, S.R.; Rice, A.M.; Rannikko, A.M.; Lentz, B.R.; Hinderliter, A. Membrane Modulates Affinity for Calcium Ion to Create an Apparent Cooperative Binding Response by Annexin A5. Biophys. J. 2013, 104, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Saurel, O.; Cezanne, L.; Milon, A.; Tocanne, J.F.; Demange, P. Influence of annexin V on the structure and dynamics of phosphatidylcholine/phosphatidylserine bilayers: A fluorescence and NMR study. Biochemistry 1998, 37, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the proteins and other materials are not available from the authors. |






| Ca2+ Titration | wtA5 | ctA5 |
|---|---|---|
| N | 4.1 ± 0.3 | ND |
| K (M−1) | 3750 ± 188 | ND |
| KD (μM) | 267 ± 13 | ND |
| ΔH (kcal/mol) | −4.03 ± 0.28 | ND |
| TΔS (kcal/mol) | 0.68 | ND |
| ΔG (kcal/mol) | −4.71 ± 0.28 | ND |
| Liposome Titration | wtA5 | ctA5 |
|---|---|---|
| N | 19.6 ± 0.4 | 18.5 ± 0.6 |
| K (M−1) | 17,900 ± 2160 | 6350 ± 614 |
| KD (μM) | 55.9 ± 6.8 | 157.5 ± 15.4 |
| ΔH (kcal/mol) | −0.42 ± 0.01 | −0.60 ± 0.02 |
| TΔS (kcal/mol) | 4.67 | 3.89 |
| ΔG (kcal/mol) | −5.09 ± 0.01 | −4.49 ± 0.02 |
| Primer | 5′ → 3′ | Target |
|---|---|---|
| e-F | GGAATTC CATATG GTGAGCAAGGGCG | EGFP |
| e-R1 | TTCCAGAACCGGTACCCTTGTACAGCTCGTCCAT | |
| e-R2 | CGC GGATCC TGAGCCACTTCCAGAACCGGTAC | |
| ea5-F | CGC GGATCC ATGGCACAGGTTCTCAGAGG | forward, wtA5 and ctA5 |
| ea5-wtR | CGTC AAGCTT ATTAGTCATCTTCTCCACAGAGC | reverse, wtA5 |
| ea5-ctR | CGTC AAGCTT ATTAGGTCTCTGCAAGGTAGGCAG | reverse, ctA5 |
| Primers | 5′ → 3′ | Target |
|---|---|---|
| i-F | GGAATTC CATATG AAAATCGAAGAAGGT | CBD-SspDnaB |
| i-R | CCG CTCGAG CCCATGGCTCTTCCGTTG | |
| ia5-F | ATCAT TGTACA CAACGCACAGGTTCTCAGAGGC | forward, wtA5 and ctA5 |
| ia5-wtR | CCG CTCGAG TTATTAGTCATCTTCTCCAC | reverse, wtA5 |
| ia5-ctR | CCG CTCGAG TTATTAGGTCTCTGCAAGGTAGGC | reverse, ctA5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, J.; Cao, Y.; Hu, M.; Hua, Z. Domain IV of Annexin A5 Is Critical for Binding Calcium and Guarantees Its Maximum Binding to the Phosphatidylserine Membrane. Molecules 2017, 22, 2256. https://doi.org/10.3390/molecules22122256
Wang J, Liu J, Cao Y, Hu M, Hua Z. Domain IV of Annexin A5 Is Critical for Binding Calcium and Guarantees Its Maximum Binding to the Phosphatidylserine Membrane. Molecules. 2017; 22(12):2256. https://doi.org/10.3390/molecules22122256
Chicago/Turabian StyleWang, Jie, Jing Liu, Yulu Cao, Minjin Hu, and Zichun Hua. 2017. "Domain IV of Annexin A5 Is Critical for Binding Calcium and Guarantees Its Maximum Binding to the Phosphatidylserine Membrane" Molecules 22, no. 12: 2256. https://doi.org/10.3390/molecules22122256




