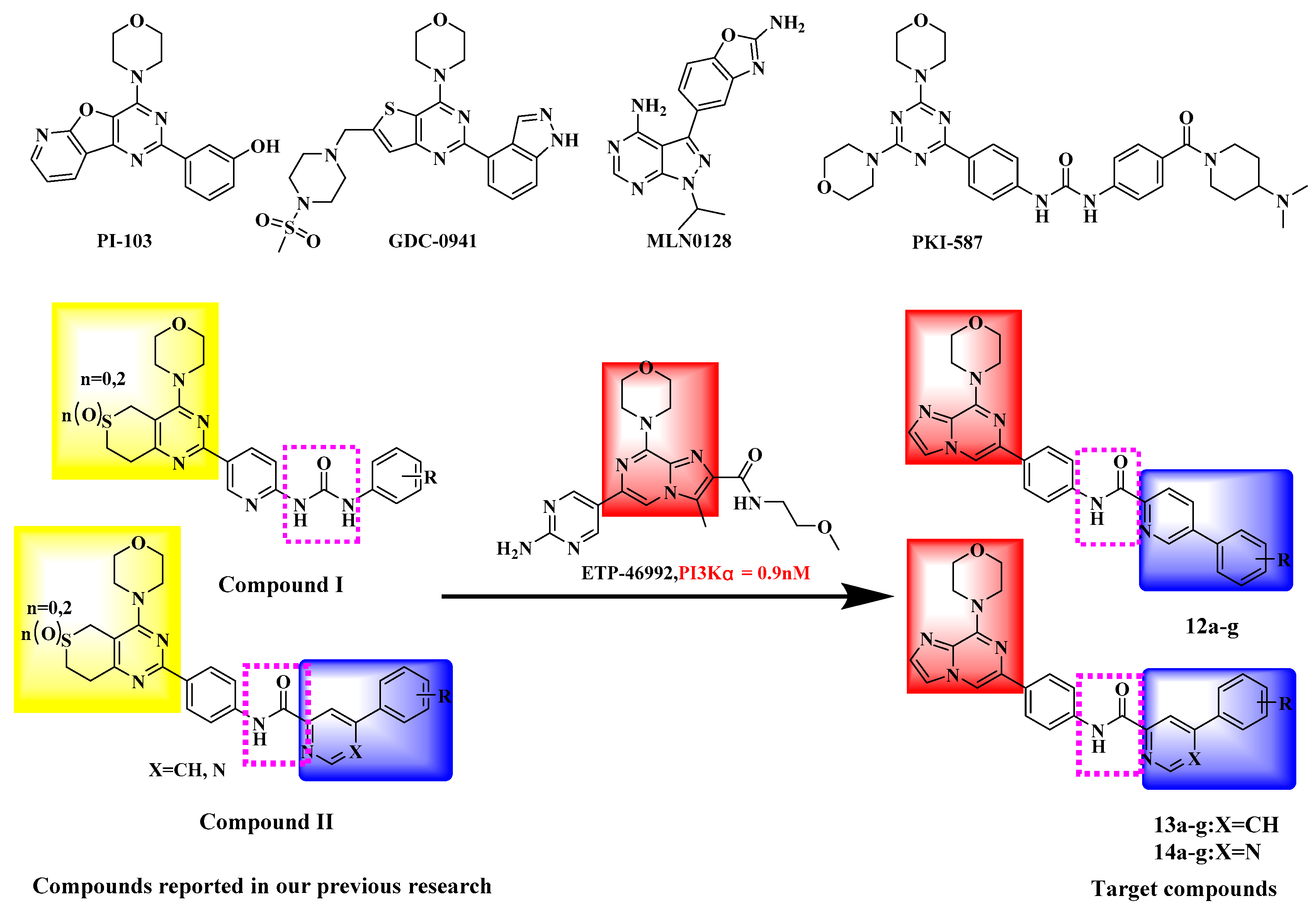
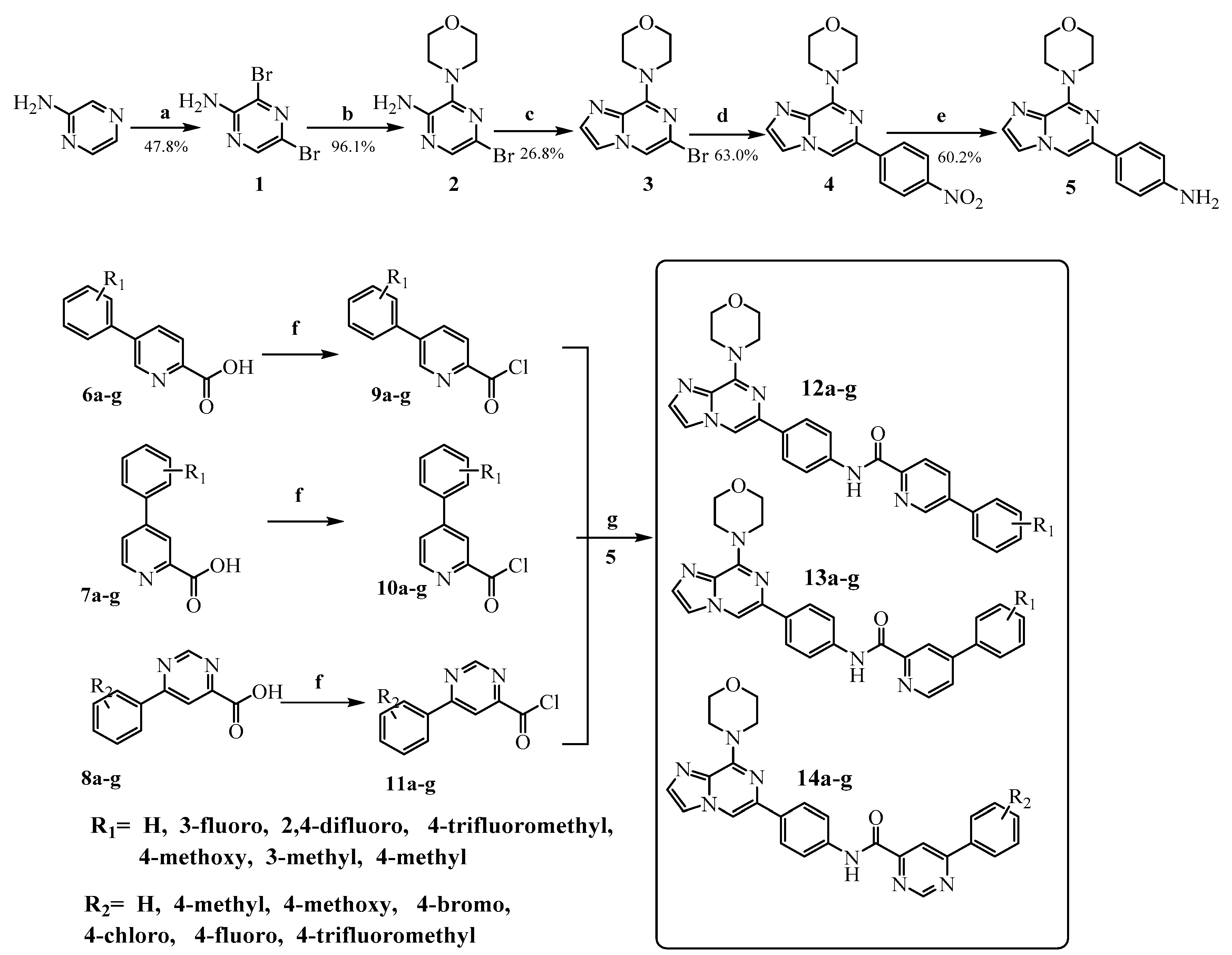
3.2. Chemistry
3,5-Dibromo-2-aminopyrazine (1)
A solution of dichloromethane (200 mL) and pyridine (25.3 mL, 0.315 mol) was added to a three-necked flask containing 2-aminopyrazine (14.27 g, 0.15 mol) and stirred well. In the dark and while refluxing a solution of bromine (16.2 mL, 0.315 mol) in dichloromethane (100 mL) was slowly added dropwise to the three-necked flask. About 1 h later the addition finished and the mixture was refluxed at 40 °C for 30 min more. After TLC monitoring indicated the reaction was complete, the reaction mixture was cooled to room temperature and distilled water (50 mL) was added and the mixture was stirred vigorously for 10 min. Then the organic layer was collected and washed twice with distilled water. Silica gel (10 g) and activated carbon (1 g) were added to the organic layer and the mixture was decolorized under reflux for 30 min. After hot filtration, the filtrate was collected and vacuum distilled. The residue was refluxed with n-hexane (45 mL) for 2 h, filtered while hot again and the solid product was dried and weighed to give 18.15 g of a pale yellow solid (47.8% yield). 1H-NMR (DMSO-d6) δ 8.14 (s, 1H), 7.01 (s, 2H).
3-Morpholino-5-bromo-2-aminopyrazine (2)
A mixture of morpholine (50 mL) and 3,5-dibromo-2-aminopyrazine (12.50 g) was refluxed for 1 h until the reaction was complete by TLC analysis. The solution was cooled to room temperature and added to a beaker containing ice water (300 mL) with continuous stirring. The solid was precipitated and filtered. After drying, 12.30 g of a yellow solid with a metallic luster was obtained (a yield of 96.1%). 1H-NMR (DMSO-d6) δ 7.70 (s, 1H), 6.28 (s, 2H), 3.85–3.62 (m, 4H), 3.04 (d, J = 4.0 Hz, 4H).
4-(6-Bromoimidazo[1,2-a]pyrazin-8-yl)morpholine (3)
Chloroacetaldehyde (4.4 g) was mixed with isopropanol (15 mL) and a portion of the mixture (10 mL) was added to a round bottom flask containing compound 2 (1.7 g). Then the temperature was raised to 45 °C and stirring was continued. After 1 h, the remaining solution was added and the temperature raised to 65 °C with continuous stirring. After TLC indicated the reaction was finished, the reaction was stopped and cooled to room temperature. Then the solution was added into a beaker containing ice water (300 mL) with continuous stirring. The brown-yellow solid that precipitated was filtered and dried to give the product in a yield of 26.8%. 1H-NMR (DMSO-d6) δ 7.93 (s, 1H), 7.25 (d, J = 7.3 Hz, 1H), 7.01 (d, J = 7.3 Hz, 1H), 3.45 (s, 4H), 3.77 (d, J = 4.3 Hz, 4H).
4-(6-(4-Nitrophenyl)imidazo[1,2-a]pyrazin-8-yl)morpholine (4)
Compound
4 was prepared using the synthetic method described in our previous article [
11]. Briefly, a solution of 1-bromo-4-nitrobenzene (2 g, 0.01 mol), bis(pinacolato) diboron (3.8 g, 0.015 mol), potassiumacetate (2.9 g, 0.03 mol) and bistriphenylphosphine palladium dichloride (0.18 g, 0.25 mmol) was added to 1,4-dioxane (50 mL) and heated to 80 °C. The reaction was continued for 3 h under N
2 and monitored by TLC. H
2O (20 mL) was added to the solution and continued for 5 min. Subsequently, compound
3 (1.7 g, 0.006 mol), Na
2CO
3 (2.1 g, 0.02 mol) and bistriphenylphosphine dichloride (0.18 g, 0.25 mmol) were added successively, then the mixture was heated to 100 °C. The reaction was continued for about 8 h. The mixture was cooled and concentrated under vacuum. H
2O (100 mL) was added, stirred for 30 min and then filtered. The filter cake was decolorized with activated carbon (5 g) and silica gel (2 g) in a mixed solvent (CH
2Cl
2:CH
3OH = 5:1, 100 mL), filtered while hot and concentrated under vacuum to obtain
4 with a yield of 63.0%. ESI-MS
m/
z: [M + H]
+ 326.1.
1H-NMR (DMSO-
d6) δ 8.78 (d,
J = 8.2 Hz 2H), 8.33 (d,
J = 8.2 Hz 2H), 8.01 (s, 1H), 7.20 (d,
J = 8.7 Hz, 1H), 6.95 (d,
J = 8.7 Hz, 1H), 3.91 (s, 4H), 3.77 (s, 4H).
4-(8-Morpholinoimidazo[1,2-a]pyridin-6-yl)aniline (5)
The synthesis of compound
5 is similar to the synthetic method reported in our previous article [
11]. Compound
4 (3.3 g, 0.01 mol) was refluxed with hydrazine hydrate (5 g, 0.1 mol), ferric chloride (0.5 g, 0.002 mol) and an appropriate amount of activated carbon in ethanol (50 mL) for 30 min–1 h and monitored by TLC. After filtering while hot and concentrating under vacuum compound
5 was obtain in a yield of 60.2%. ESI-MS
m/
z: [M + H]
+ 296.2. [M + H]
+ 326.1.
1H-NMR (DMSO-
d6) δ 8.12 (d,
J = 8.5 Hz 2H), 7.89 (s, 1H), 7.12 (d,
J = 8.3 Hz, 1H), 6.99 (d,
J = 8.3 Hz, 1H), 6.60 (d,
J =8.5 Hz 2H), 5.56 (s, 2H), 3.75 (d,
J = 3.5 Hz, 4H), 3.32 (s, 4H).
General Procedure for the Preparation of Compounds 6a–g, 7a–g, 8a–g, 9a–g, 10a–g and 11a–g
Compounds
6a–
g,
7a–
g and
8a–
g were synthesized according to the procedures reported by our research group [
11,
12]. Then compounds
6a–
g,
7a–
g and
8a–
g (0.02 mmol) were dissolved in thionyl chloride (8 mL) and refluxed for 1 h. The reaction mixture was evaporated to yield the corresponding chloride which was dissolved in dichloromethane (10 mL). The solution of
9a–
g,
10a–
g or
11a–
g was used for the next step without further purification.
General Procedure for the Preparation of the Target Compounds 12a–g, 13a–g and 14a–g
A solution of phenylpyrimidine chloride 9a–g, 10a–g or 11a–g in dichloromethane (10 mL) was added dropwise to a solution of 5 (0.12 mol) and N,N-diisopropylethylamine (0.048 mol) in dichloromethane (10 mL) in an ice bath. Upon completion of the addition, the reaction mixture was removed from the ice bath, held at room temperature for 15 min and monitored by thin-layer chromatography (TLC). The mixture was washed with 10% K2CO3 (50 mL × 3) followed by brine (50 mL × 1), and the organic layer was separated, dried over anhydrous sodium sulfate, and evaporated to yield the target compounds 12a–g, 13a–g and 14a–g which were recrystallized from isopropanol. In addition, all the target compounds were purified by silica gel column chromatography (CH2Cl2:CH3OH = 100:1).
N-(4-Morpholinoimidazo[1,2-a]pyridin-6-yl) phenyl)-4-phenylpyridine amide (12a). A yellow solid; yield: 96.1%; m.p. 268–270 °C; ESI-MS [M + H]+ m/z: 477.2; 1H-NMR (CDCl3) δ 10.46 (s, 1H), 9.06 (s, 1H), 8.86 (s, 2H), 8.39 (d, J = 8.0 Hz, 1H), 8.28 (d, J = 8.1 Hz, 1H), 8.22 (d, J = 8.2 Hz, 1H), 8.13–8.04 (m, 4H), 7.96 (s, 3H), 7.83 (s, 2H), 4.21 (s, 4H), 4.01 (s, 4H).
5-(3-Fluorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (12b). A yellow solid; yield: 79.4%; m.p. 261–264 °C; ESI-MS [M + H]+ m/z: 495.2; 1H-NMR (400 MHz, CDCl3) δ 10.18 (s, 1H), 8.85 (s, 2H), 8.40 (d, J = 8.1 Hz, 2H), 8.29 (d, J = 8.1 Hz, 1H), 8.09 (s, 3H), 7.88 (s, 1H), 7.75 (s, 1H), 7.52 (d, J = 5.1 Hz, 1H), 7.45 (s, 1H), 7.36 (d, J = 9.2 Hz, 1H), 7.20 (d, J = 9.0 Hz, 1H), 4.17 (s, 4H), 4.03 (s, 4H).
5-(2,4-Difluorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (12c). A pale yellow solid; yield: 90.2%; m.p. 255–257 °C; ESI-MS [M + H]+ m/z: 513.2; 1H-NMR (DMSO-d6) δ 10.97–10.71 (m, 1H), 8.86 (s, 1H), 8.35 (d, J = 9.2 Hz, 1H), 8.22 (s, 1H), 8.04 (s, 1H), 7.84–7.71 (m, 1H), 7.60 (d, J = 12.2 Hz, 2H), 7.51–7.37 (m, 1H), 7.32–7.17 (m, 1H), 6.51 (s, 2H), 6.30 (s, 2H), 3.92 (s, 4H), 3.74 (s, 4H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-5-(4-(trifluoromethyl)phenyl)picolinamide (12d). A gray solid; yield: 83.5%; m.p. 233–235 °C; ESI-MS [M + H]+ m/z: 545.2; 1H-NMR (CDCl3) δ 10.46 (s, 1H), 8.88 (s, 2H), 8.33 (d, J = 8.0 Hz, 2H), 8.23 (d, J = 14.9 Hz, 1H), 8.12 (s, 2H), 7.99 (d, J = 22.6 Hz, 5H), 7.56 (s, 1H), 7.48 (s, 1H), 4.08 (d, J = 18.4 Hz, 8H).13C-NMR (DMSO-d6) δ 162.66, 157.43, 149.99, 147.87, 147.28, 138.75, 137.36, 136.99, 136.53, 135.92, 132.97, 132.96, 132.52, 132.29, 128.69 (2C), 128.68 (2C), 128.52, 126.28 (2C), 123.14, 120.76 (2C), 116.11, 66.73 (2C),46.71 (2C).
5-(4-Methoxyphenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (12e). A pale yellow solid; yield: 93.3%; m.p. 255–257 °C; ESI-MS [M + H]+ m/z: 507.2; 1H-NMR (DMSO-d6) δ 11.05 (s, 1H), 8.74 (d, J = 5.1 Hz, 1H), 8.57 (s, 1H), 8.47 (s, 1H), 8.03 (d, J = 8.6 Hz, 2H), 8.02 (s, 2H), 7.87 (d, J = 9.8 Hz, 2H), 7.86 (d, J = 8.6 Hz, 2H), 7.51 (s, 1H), 7.14 (d, J = 8.6 Hz, 2H), 4.32 (s, 4H), 4.19 (s, 2H), 3.85 (d, J = 3.9 Hz, 3H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-5-(m-tolyl)picolinamide (12f). A pale yellow solid; yield: 72.6%; m.p. 241–243 °C; ESI-MS [M + H]+ m/z: 491.2; 1H-NMR (CDCl3) δ 10.17 (s, 1H), 8.84 (s, 1H), 8.36 (d, J = 8.0 Hz, 1H), 8.30–8.22 (m, 1H), 8.20 (d, J = 7.5 Hz, 1H), 8.10 (d, J = 9.8 Hz, 2H), 8.06 (s, 2H), 7.93 (d, J = 8.2 Hz, 2H), 7.74 (s, 1H), 7.56 (d, J = 7.3 Hz, 3H), 4.25 (s, 4H), 3.98 (s, 4H), 2.44 (s, 3H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-5-(p-tolyl)picolinamide (12g). A pale yellow solid; yield: 89.1%; m.p. 258–260 °C; ESI-MS [M + H]+ m/z: 491.2; 1H-NMR (DMSO-d6) δ 10.79 (s, 1H), 9.02 (d, J = 9.4 Hz, 2H), 8.59 (s, 1H), 8.35 (d, J = 8.1 Hz, 1H), 8.24 (d, J = 8.1 Hz, 1H), 8.05 (d, J = 8.7 Hz, 2H), 7.77 (s, 2H), 7.59 (s, 2H), 7.37 (t, J = 8.5 Hz, 3H), 4.30 (s, 4H), 3.80 (s, 4H), 2.40 (s, 3H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-4-phenylpicolinamide (13a). A yellow solid; yield: 97.0%; m.p. 261–262 °C; ESI-MS [M + H]+ m/z: 477.2; 1H-NMR (DMSO-d6) δ 9.55 (s, 1H), 7.59 (d, J = 5.0 Hz, 1H), 7.34 (s, 1H), 7.20 (s, 1H), 6.79 (d, J = 7.8 Hz, 5H), 6.74–6.65 (m, 3H), 6.34 (dd, J = 8.4, 4.3 Hz, 4H), 3.07 (s, 4H), 2.61–2.51 (m, 4H).
4-(3-Fluorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (13b). A white solid; yield: 88.0%; m.p. 237–239 °C; ESI-MS [M + H]+ m/z: 495.2; 1H-NMR (DMSO-d6) δ 10.85 (s, 1H), 8.84 (d, J = 5.0 Hz, 1H), 8.80 (d, J = 5.0 Hz, 1H), 8.60 (s, 1H), 8.45 (s, 1H), 8.33 (s, 1H), 8.06 (dd, J = 5.7, 3.5 Hz, 3H), 7.96 (s, 1H), 7.85–7.79 (m, 2H), 7.63 (d, J = 7.7 Hz, 1H), 7.45–7.33 (m, 2H), 4.30 (s, 4H), 3.80 (d, J = 4.2 Hz, 4H).
4-(2,4-Difluorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (13c). A pale yellow solid; yield: 91.2%; m.p. 266–267 °C; ESI-MS [M + H]+ m/z: 513.2; 1H-NMR (DMSO-d6) δ 10.76 (s, 1H), 8.79 (d, J = 5.1 Hz, 1H), 8.52 (s, 1H), 8.26 (s, 1H), 7.96 (d, J = 7.3 Hz, 4H), 7.89 (s, 1H), 7.85–7.74 (m, 2H), 7.52 (s, 1H), 7.43 (d, J = 9.6 Hz, 1H), 7.26 (t, J = 8.3 Hz, 1H), 4.23 (s, 4H), 3.74 (d, J = 4.3 Hz, 4H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-4-(4-(trifluoromethyl)phenyl)picolinamide (13d). A pale yellow solid; yield: 81.8%; m.p. 269–272 °C; ESI-MS [M + H]+ m/z: 545.2; 1H-NMR (DMSO-d6) δ 10.86 (s, 1H), 8.88 (d, J = 5.0 Hz, 1H), 8.59 (s, 1H), 8.49 (s, 1H), 8.15 (d, J = 8.0 Hz, 2H), 8.13–8.07 (m, 2H), 8.03 (d, J = 11.5 Hz, 3H), 8.00 (s, 1H), 7.95 (d, J = 8.4 Hz, 2H), 7.59 (s, 1H), 4.30 (s, 4H), 3.80 (d, J = 4.0 Hz, 4H).
4-(4-Methoxyphenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)picolinamide (13e). A pale yellow solid; yield: 93.1%; m.p. 257–260 °C; ESI-MS [M + H]+ m/z: 507.2; 1H-NMR (DMSO-d6) δ 10.82 (s, 1H), 8.76 (d, J = 5.1 Hz, 1H), 8.60 (s, 1H), 8.40 (s, 1H), 8.05 (d, J = 8.6 Hz, 2H), 8.02 (s, 2H), 7.97 (d, J = 9.8 Hz, 2H), 7.91 (d, J = 8.6 Hz, 2H), 7.59 (s, 1H), 7.14 (d, J = 8.6 Hz, 2H), 4.30 (s, 4H), 3.85 (s, 2H), 3.80 (d, J = 3.9 Hz, 3H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-4-(m-tolyl)picolinamide (13f). A pale red solid; yield: 91.5%; m.p. 233–234 °C; ESI-MS [M + H]+ m/z: 491.2; 1H-NMR (DMSO-d6) δ 10.82 (s, 1H), 8.79 (d, J = 4.9 Hz, 1H), 8.59 (s, 1H), 8.42 (s, 1H), 8.30 (s, 1H), 8.05 (d, J = 8.4 Hz, 3H), 7.83 (d, J = 8.1 Hz, 3H), 7.59 (s, 1H), 7.43–7.36 (m, 3H), 4.30 (s, 4H), 3.80 (d, J = 3.7 Hz, 4H), 2.40 (s, 3H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-4-(p-tolyl)picolinamide (13g). A brown solid; yield: 98.0%; m.p. 271–273 °C; ESI-MS [M + H]+ m/z: 491.2; 1H-NMR (DMSO-d6) δ 9.87 (s, 1H), 7.90 (d, J = 5.2 Hz, 1H), 7.56 (s, 1H), 7.44 (s, 1H), 7.22–7.15 (m, 3H), 6.95 (d, J = 8.5 Hz, 3H), 6.73 (s, 1H), 6.54 (s, 4H), 3.45 (s, 4H), 3.15–2.68 (m, 4H), 2.33 (s, 3H).13C-NMR (DMSO-d6) δ 162.98, 151.15, 149.90, 149.64, 149.31, 147.87, 140.08, 138.78, 135.91, 134.03, 132.94, 132.30, 130.48 (2C), 127.35 (2C), 127.31, 126.29 (2C), 120.70, 119.64 (2C), 116.13, 116.11, 66.73 (2C), 46.70 (2C), 21.29.
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-6-phenylpyrimidine-4-carboxamide (14a). A pale yellow solid; yield: 82.7%; m.p. 272–275 °C; ESI-MS [M + H]+ m/z: 478.1; 1H-NMR (DMSO-d6) δ 10.79 (s, 1H), 8.75 (d, J = 5.1 Hz, 1H), 8.58 (s, 1H), 8.39 (s, 1H), 8.04 (d, J = 8.7 Hz, 2H), 8.01 (s, 1H), 7.98 (d, J = 6.3 Hz, 2H), 7.95 (s, 1H), 7.89 (d, J = 8.5 Hz, 2H), 7.58 (s, 1H), 7.13 (d, J = 8.5 Hz, 2H), 4.29 (s, 4H), 3.80 (d, J = 4.3 Hz, 4H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-6-(p-tolyl)pyrimidine-4-carboxamide (14b). A pale yellow solid; yield: 79.5%; m.p. 222–225 °C; ESI-MS [M + H]+ m/z: 492.2; 1H-NMR (DMSO-d6) δ 10.82 (s, 1H), 8.77 (dd, J = 22.5, 5.1 Hz, 2H), 8.58 (s, 1H), 8.41 (s, 1H), 8.27 (s, 1H), 7.94–7.87 (m, 2H), 7.58 (s, 2H), 7.41 (t, J = 8.3 Hz, 4H), 4.29 (s, 4H), 3.79 (s, 4H), 1.25 (d, J = 6.5 Hz, 3H).
6-(4-Methoxyphenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)pyrimidine-4-carboxamide (14c). A pale brown solid; yield: 77.8%; m.p. 269–271 °C; ESI-MS [M + H]+ m/z: 508.2; 1H-NMR (DMSO-d6) δ 10.96 (s, 1H), 9.38 (s, 1H), 8.57 (s, 1H), 8.32 (d, J = 8.0 Hz, 2H), 8.03 (dd, J = 14.7, 8.4 Hz, 4H), 7.96 (s, 1H), 7.08 (d, J = 1.7 Hz, 2H), 7.05 (d, J = 1.6 Hz, 2H), 4.29 (s, 4H), 3.87 (s, 3H), 3.80 (s, 4H). 13C-NMR (DMSO-d6) δ 162.80, 162.53, 162.06, 158.73, 158.48, 157.98, 157.00, 138.37, 133.36, 132.96, 132.29, 129.59 (2C), 128.25, 128.20, 126.30 (2C), 120.98 (2C), 116.12, 114.98 (3C), 66.73 (2C), 55.95, 46.70 (2C).
6-(4-Bromophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)pyrimidine-4-carboxamide (14d). A white solid; yield: 90.9%; m.p. 221–223 °C; ESI-MS [M + H]+ m/z: 557.1; 1H-NMR (DMSO-d6) δ 10.91 (s, 1H), 9.49 (s, 1H), 8.65 (s, 1H), 8.59 (d, J = 11.9 Hz, 2H), 8.38 (d, J = 8.3 Hz, 2H), 8.00–7.94 (m, 3H), 7.85 (d, J = 8.3 Hz, 2H), 7.59 (d, J = 2.7 Hz, 2H), 4.30 (s, 4H), 3.79 (d, J = 4.0 Hz, 4H).
6-(4-Chlorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)pyrimidine-4-carboxamide (14e). A pale yellow solid; yield: 93.2%; m.p. 235–237 °C; ESI-MS [M + H]+ m/z: 513. 1; 1H-NMR (DMSO-d6) δ 10.81 (s, 1H), 8.78 (d, J = 5.1 Hz, 1H), 8.58 (s, 1H), 8.41 (s, 1H), 8.05 (d, J = 8.8 Hz, 2H), 8.00 (d, J = 7.5 Hz, 2H), 7.95 (s, 1H), 7.83 (d, J = 8.0 Hz, 2H), 7.58 (s, 1H), 7.41 (d, J = 8.0 Hz, 2H), 4.29 (s, 4H), 3.88–3.67 (m, 4H).
6-(4-Fluorophenyl)-N-(4-(8-morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)pyrimidine-4-carboxamide (14f). A gray solid; yield: 80.8%; m.p. 259–260 °C; ESI-MS [M + H]+ m/z: 496.2; 1H-NMR (CDCl3) δ 10.05 (s, 1H), 9.33 (s, 1H), 8.97 (s, 1H), 8.63 (s, 1H), 8.17 (dd, J = 18.2, 8.6 Hz, 2H), 8.10–7.83 (m, 5H), 7.75 (d, J = 5.2 Hz, 1H), 7.55 (d, J = 8.0 Hz, 2H), 4.32 (s, 4H), 3.96 (s, 4H).
N-(4-(8-Morpholinoimidazo[1,2-a]pyrazin-6-yl)phenyl)-6-(4-(trifluoromethyl)phenyl)pyrimidine-4-carboxamide (14g). A white solid; yield: 77.6%; m.p. 278–281 °C; ESI-MS [M + H]+ m/z: 546.2; 1H-NMR (DMSO-d6) δ 10.86 (s, 1H), 9.86 (d, J = 5.0 Hz, 1H), 8.49 (s, 1H), 8.15 (d, J = 8.0 Hz, 2H), 8.10 (d, J = 4.5 Hz, 1H), 8.03 (dd, J = 20.1, 8.6 Hz, 4H), 7.95 (d, J = 8.4 Hz, 3H), 7.59 (s, 1H), 4.30 (s, 4H), 3.80 (s, 4H).