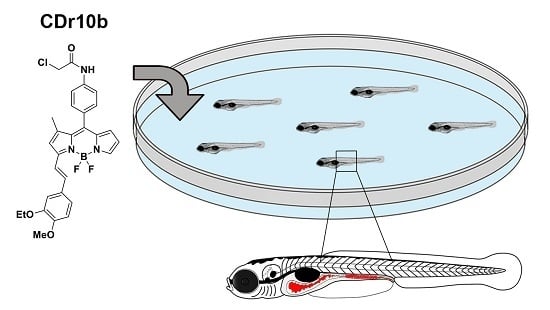
The Vital Dye CDr10b Labels the Zebrafish Mid-Intestine and Lumen
Abstract
:1. Introduction
2. Results and Discussion
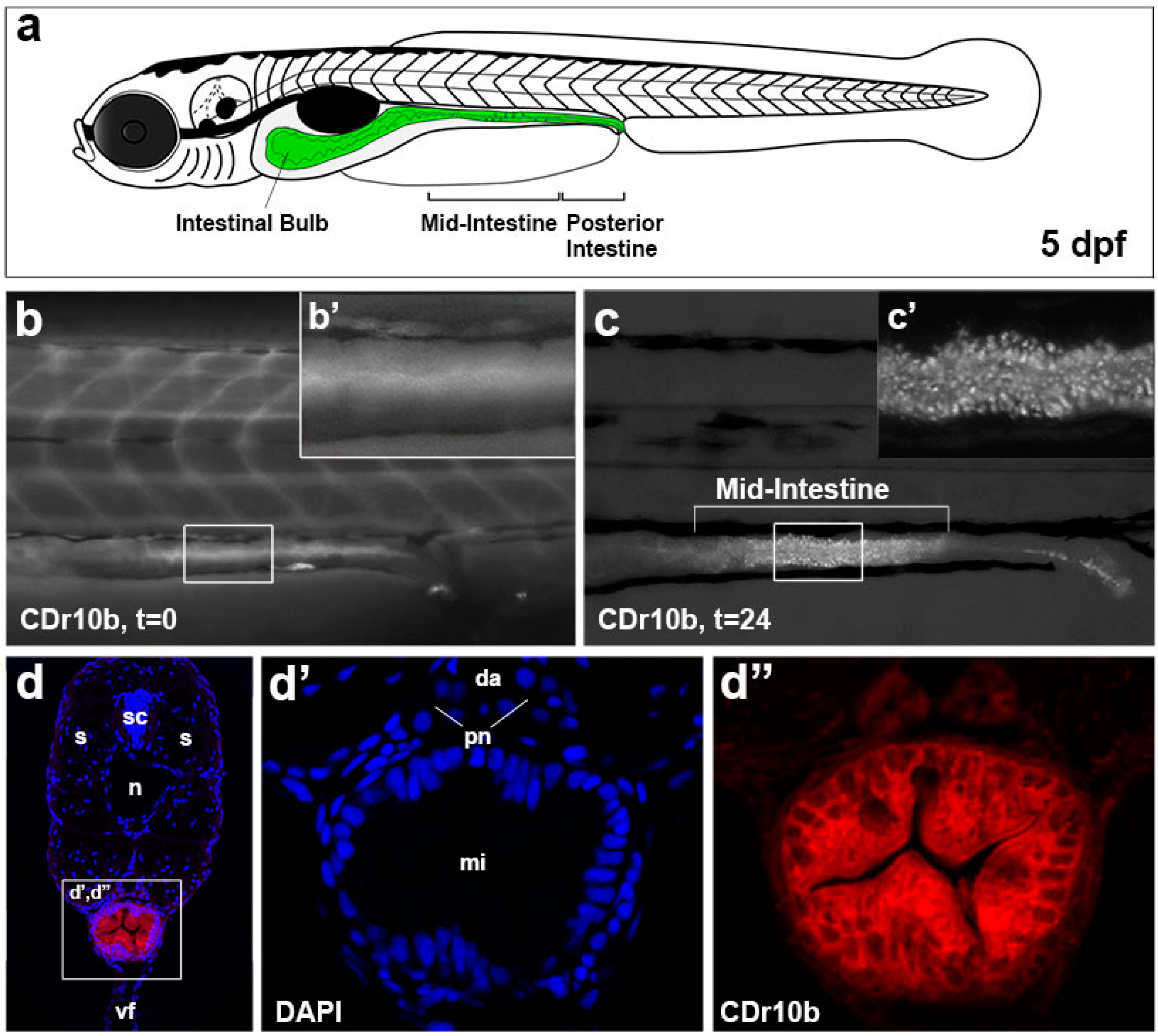
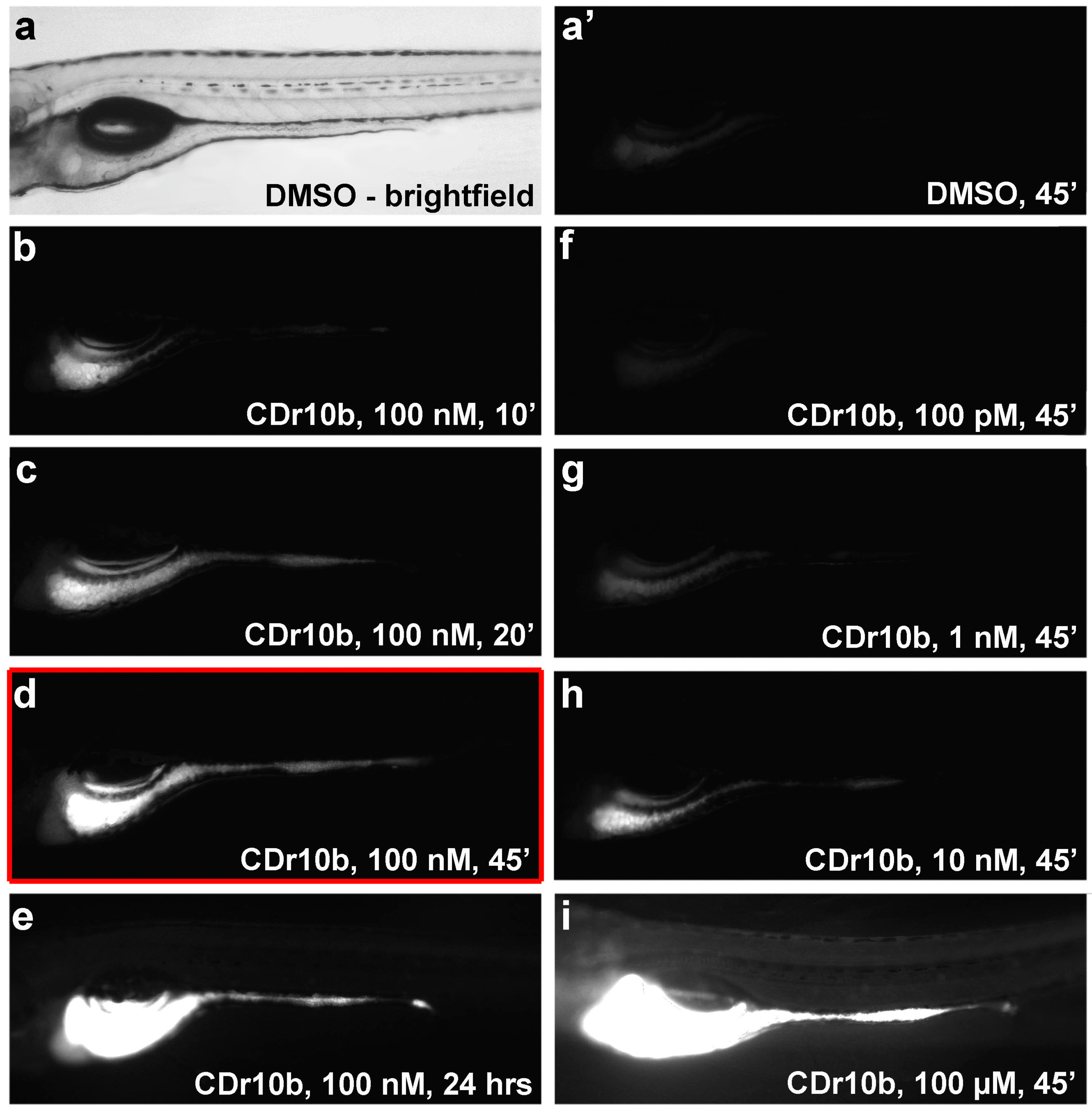
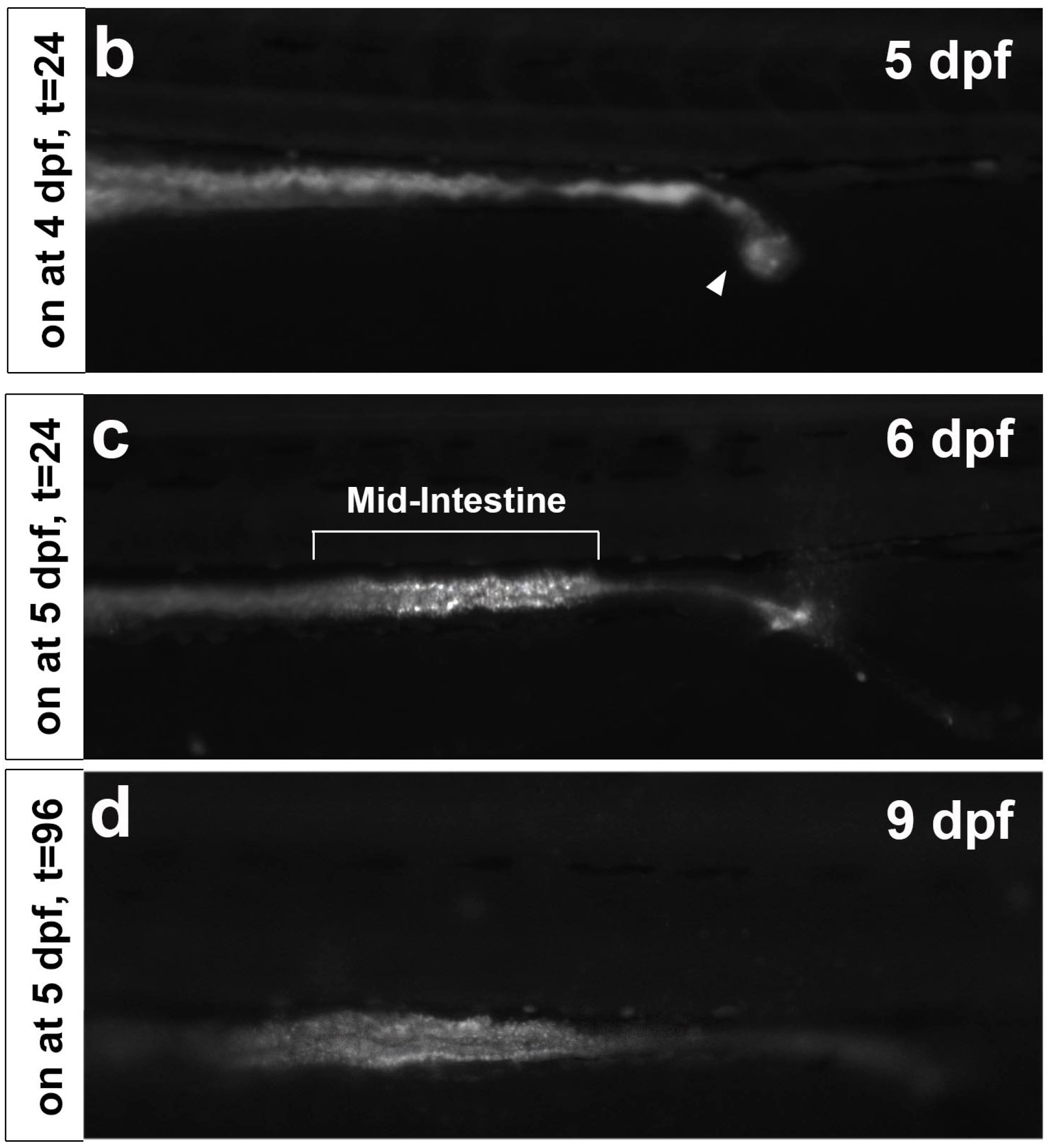
2.1. CDr10b Labels the Larval Mid-Intestine
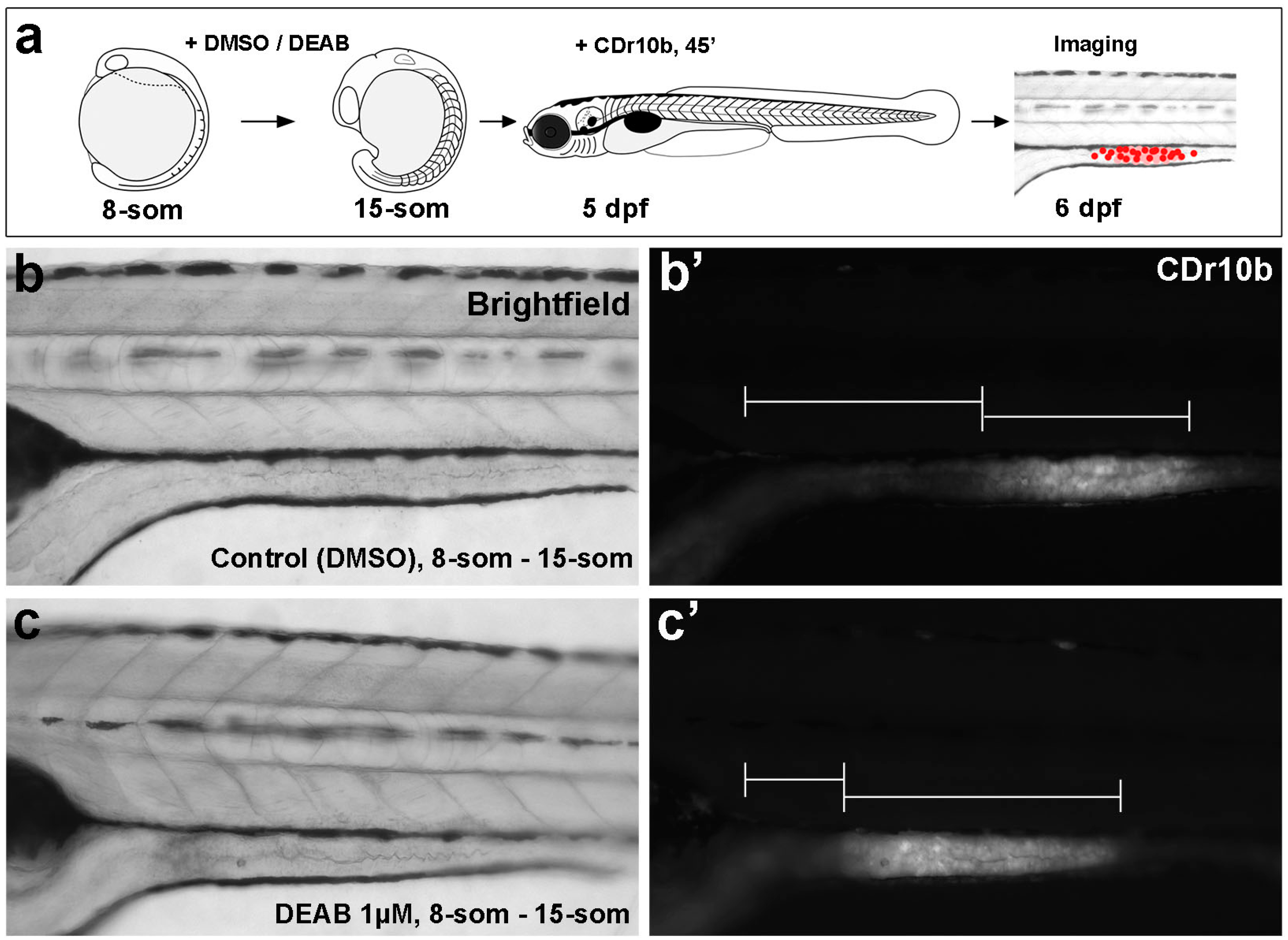
2.2. CDr10b Allows Detection of Intestinal Patterning Defects
2.3. CDr10b as a Readout for Intestinal Integrity
3. Materials and Methods
3.1. Synthesis and Characterization of CDr10b
3.2. CDr10b/CDr10a
3.3. Zebrafish Maintenance and Stocks
3.4. Zebrafish Treatment
3.5. Imaging
3.6. Duration, Dose and Toxicity
3.7. Histology
3.8. Laser Ablation
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Maloy, K.J.; Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011, 474, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Sander, V.; Davidson, A.J. Kidney injury and regeneration in zebrafish. Semin. Nephrol. 2014, 34, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Du, J.; Lam, S.H.; Mathavan, S.; Matsudaira, P.; Gong, Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genom. 2010, 11, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, J.K. Transcriptional networks and signaling pathways that govern vertebrate intestinal development. Curr. Top. Dev. Biol. 2010, 90, 159–192. [Google Scholar] [PubMed]
- Ng, A.N.Y.; de Jong-Curtain, T.A.; Mawdsley, D.J.; White, S.J.; Shin, J.; Appel, B.; Dong, P.D.S.; Stainier, D.Y.R.; Heath, J.K. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev. Biol. 2005, 286, 114–135. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.N.; Akhter, S.; Smith, E.M.; Lorent, K.; Pack, M. Intestinal growth and differentiation in zebrafish. Mech. Dev. 2005, 122, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Samarut, E.; Fraher, D.; Laudet, V.; Gibert, Y. ZebRA: An overview of retinoic acid signaling during zebrafish development. Biochim. Biophys. Acta 2015, 1849, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Stafford, D.; Prince, V.E. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 2002, 12, 1215–1220. [Google Scholar] [CrossRef]
- Kelly, G.M.; Drysdale, T.A. Retinoic acid and the development of the endoderm. J. Dev. Biol. 2015, 3, 25–56. [Google Scholar] [CrossRef]
- Sturm, A.; Dignass, A.U. Epithelial restitution and wound healing in inflammatory bowel disease. World J. Gastroenterol. 2008, 14, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, M.; Konno, S. Wound healing of intestinal epithelial cells. World J. Gastroenterol. 2011, 17, 2161–2171. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Jankowski, J.; Goldsmith, P. In vivo analysis of gut function and disease changes in a zebrafish larvae model of inflammatory bowel disease: A feasibility study. Inflamm. Bowel Dis. 2010, 16, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Oehlers, S.H.; Flores, M.V.; Okuda, K.S.; Hall, C.J.; Crosier, K.E.; Crosier, P.S. A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 2011, 240, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, J.R.; Cocchiaro, J.L.; Rawls, J.F.; Jobin, C. Glafenine-induced intestinal injury in zebrafish is ameliorated by μ-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis. Model Mech. 2013, 6, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Cocchiaro, J.L.; Rawls, J.F. Microgavage of zebrafish larvae. J. Vis. Exp. 2013, 72, e4434. [Google Scholar] [CrossRef] [PubMed]
- Carten, J.D.; Bradford, M.K.; Farber, S.A. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 2011, 360, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Sander, V.; Patke, S.; Sahu, S.; Teoh, C.L.; Peng, Z.; Chang, Y.-T.; Davidson, A.J. The small molecule probe PT-Yellow labels the renal proximal tubules in zebrafish. Chem. Commun. 2014, 51, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, Y.K.; Vendrell, M.; Chang, Y.-T. Diversity-oriented fluorescence library approach for the discovery of sensors and probes. Mol. BioSyst. 2009, 5, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Leong, C.; Lee, S.C.; Ock, J.; Li, X.; See, P.; Park, S.J.; Ginhoux, F.; Yun, S.-W.; Chang, Y.-T. Microglia specific fluorescent probes for live cell imaging. Chem. Commun. 2014, 50, 1089–1091. [Google Scholar] [CrossRef] [PubMed]
- Nadauld, L.D.; Sandoval, I.T.; Chidester, S.; Yost, H.J.; Jones, D.A. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J. Biol. Chem. 2004, 279, 51581–51589. [Google Scholar] [CrossRef] [PubMed]
- Nadauld, L.D.; Shelton, D.N.; Chidester, S.; Yost, H.J.; Jones, D.A. The zebrafish retinol dehydrogenase, rdh1l, is essential for intestinal development and is regulated by the tumor suppressor adenomatous polyposis coli. J. Biol. Chem. 2005, 280, 30490–30495. [Google Scholar] [CrossRef] [PubMed]
- Wingert, R.A.; Selleck, R.; Yu, J.; Song, H.D.; Chen, Z.; Song, A.; Zhou, Y.; Thisse, B.; Thisse, C.; McMahon, A.P.; Davidson, A.J. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007, 3, 1922–1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- McLeish, J.A.; Chico, T.J.; Taylor, H.B.; Tucker, C.; Donaldson, K.; Brown, S.B. Skin exposure to micro- and nano-particles can cause haemostasis in zebrafish larvae. Thromb Haemost. 2010, 103, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, J.R.; Tomkovich, S.; Jobin, C. A Rapid Screenable Assay for Compounds That Protect Against Intestinal Injury in Zebrafish Larva. Methods Mol. Biol. 2016, 1422, 281–293. [Google Scholar] [PubMed]
- Sample Availability: Limited quantities of compound CDr10b are available from the authors.


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sander, V.; Patke, S.; Lee, J.Y.; Chang, Y.-T.; Davidson, A.J. The Vital Dye CDr10b Labels the Zebrafish Mid-Intestine and Lumen. Molecules 2017, 22, 454. https://doi.org/10.3390/molecules22030454
Sander V, Patke S, Lee JY, Chang Y-T, Davidson AJ. The Vital Dye CDr10b Labels the Zebrafish Mid-Intestine and Lumen. Molecules. 2017; 22(3):454. https://doi.org/10.3390/molecules22030454
Chicago/Turabian StyleSander, Veronika, Shantanu Patke, Jung Y. Lee, Young-Tae Chang, and Alan J. Davidson. 2017. "The Vital Dye CDr10b Labels the Zebrafish Mid-Intestine and Lumen" Molecules 22, no. 3: 454. https://doi.org/10.3390/molecules22030454