Strengthening Triterpene Saponins Biosynthesis by Over-Expression of Farnesyl Pyrophosphate Synthase Gene and RNA Interference of Cycloartenol Synthase Gene in Panax notoginseng Cells
Abstract
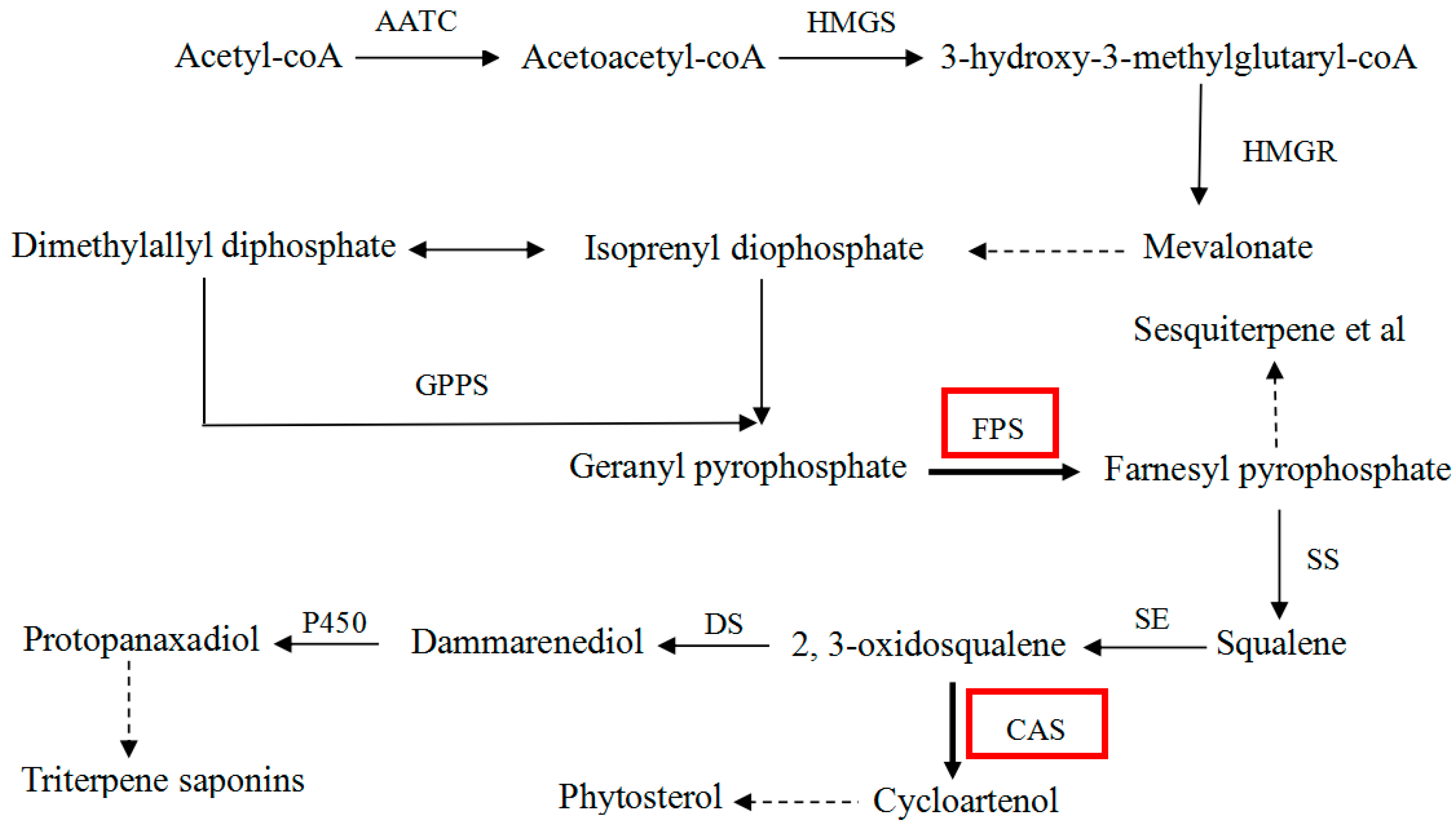
:1. Introduction
2. Results and Discussion
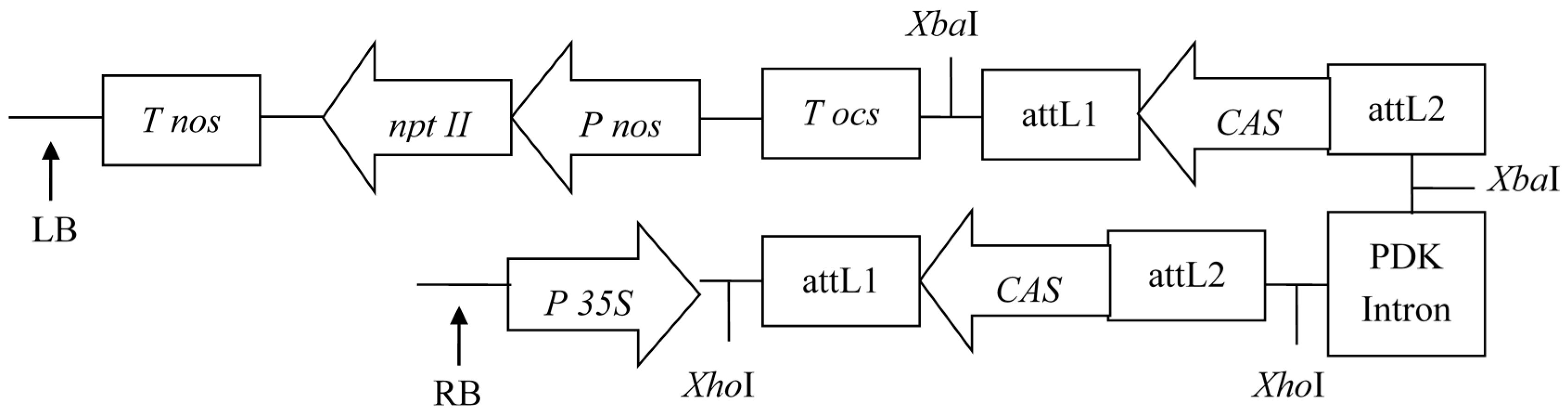
2.1. RNAi and Over-Expression Vector Construction
2.2. Genetic Transformation of P. notoginseng
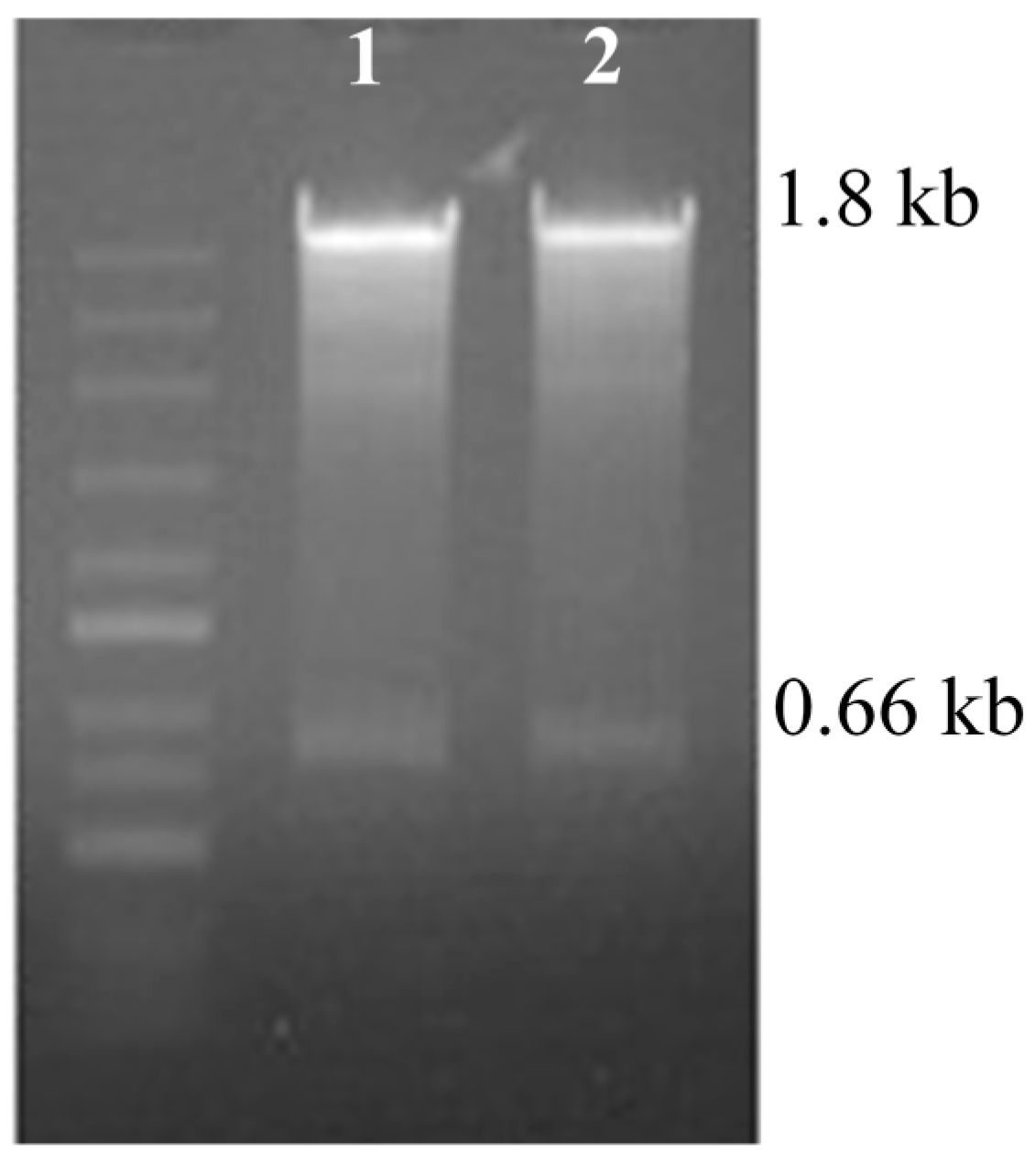
2.3. PCR Analysis of Neomycin Phosphotransferase II (nptII) Gene and Hygromycin Phosphotransferase II (hptII) Gene
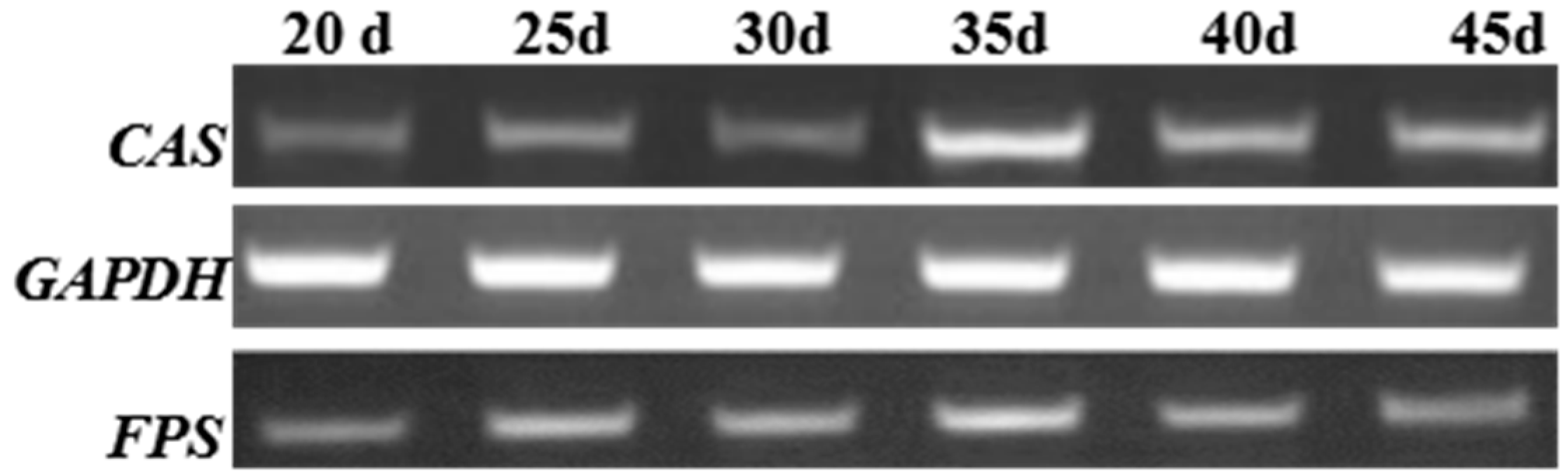
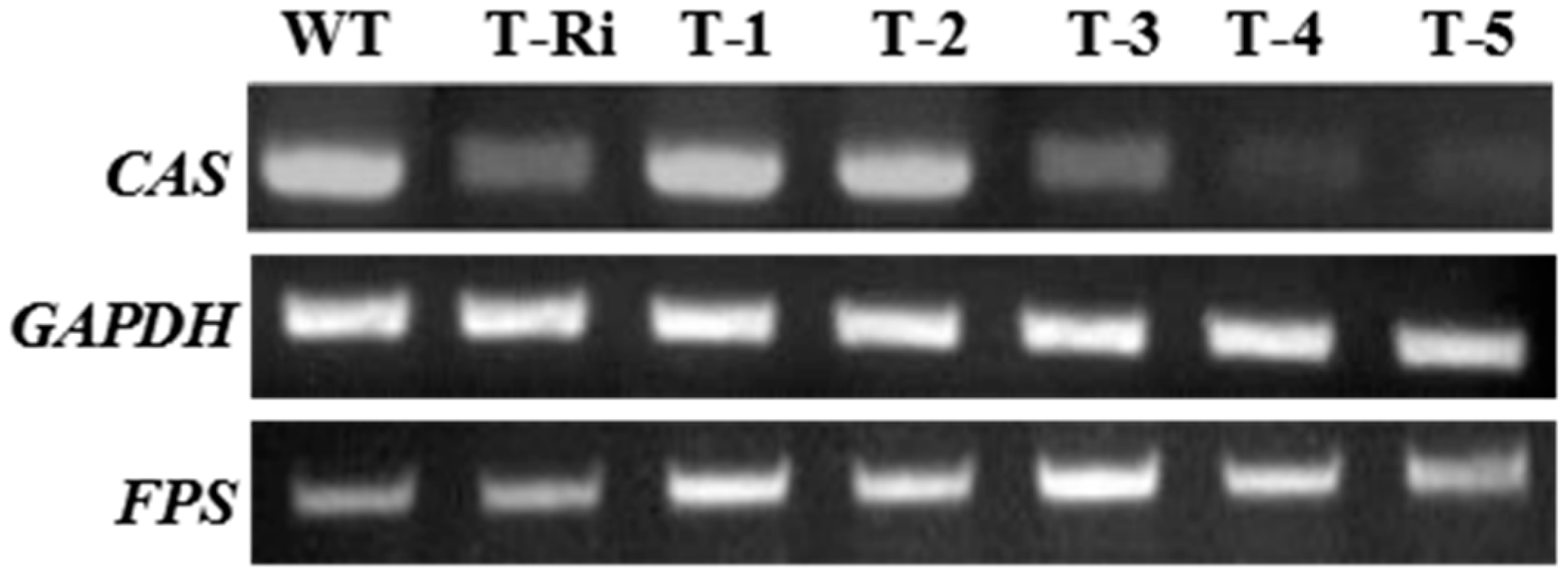
2.4. Reverse-Transcriptase-PCR(RT-PCR) Analysis
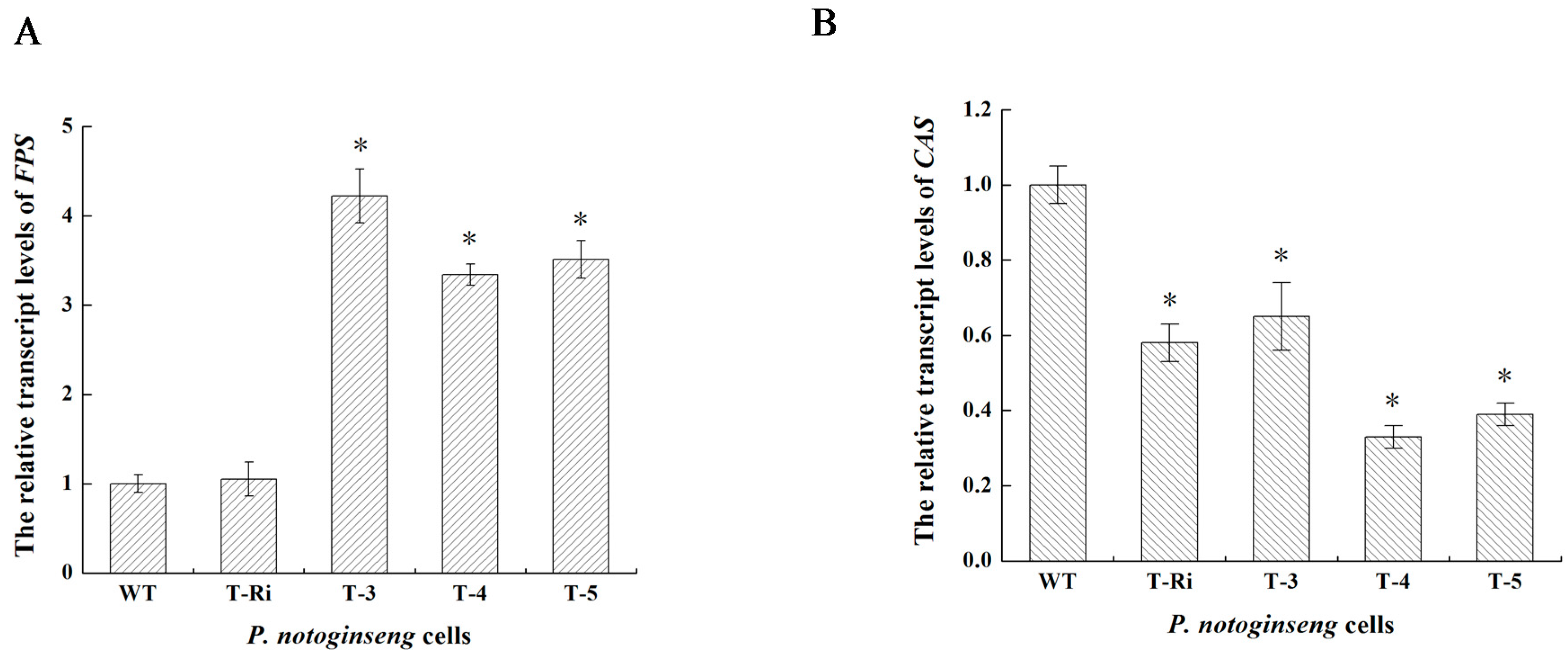
2.5. qPCR Analysis
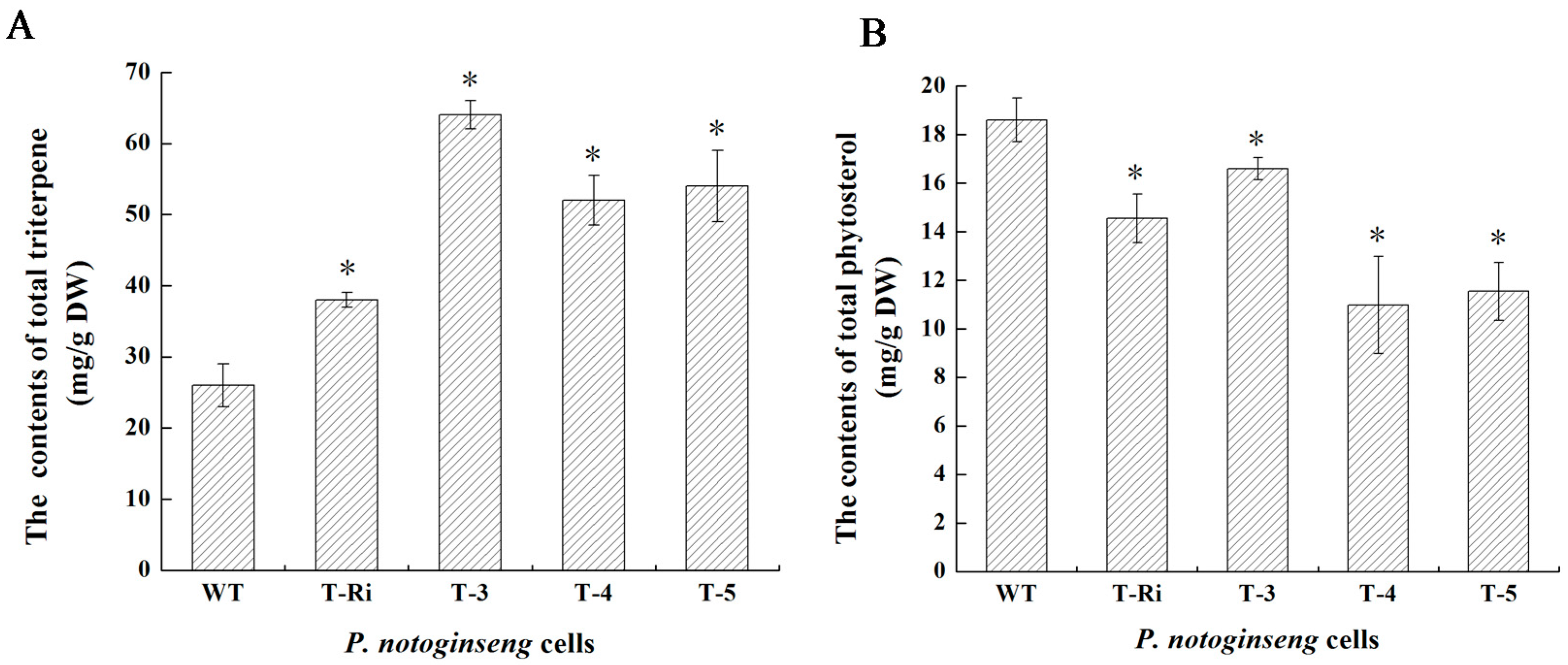
2.6. Metabolic Analysis
3. Materials and Methods
3.1. Plant Materials
3.2. RNA Isolation and cDNA Synthesis
3.3. RNAi Vector of CAS Construction
3.4. Over-Expression Vector of FPS Construction
3.5. Genetic Transformation of P. notoginseng Cells
3.6. PCR Analysis of nptII and hptII Genes
3.7. Semiquantitative RT-PCR Analysis
3.8. Expression Analysis of FPS and CAS by Quantitative Real-Time PCR (qPCR)
3.9. Quantitative Analysis of Total Triterpene Saponins
3.10. High-Performance Liquid Chromatography (HPLC) Analysis of Four Major Ginsenosides
3.11. Quantitative Analysis of Totalphytosterols
3.12. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Niu, Y.Y.; Luo, H.M.; Sun, C.; Yang, T.J.; Dong, L.L.; Huang, L.F.; Chen, S.L. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene 2014, 533, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Li, Y.; Li, S.L.; Yang, J.; Zhang, P.H.; Wang, Q. Chemical profiles and anticancer effects of saponin fractions of different polarity from the leaves of Panax notoginseng. Chin. J. Nat. Med. 2014, 12, 30–37. [Google Scholar]
- Liu, J.J.; Wang, Y.T.; Qiu, L.; Yu, Y.Y.; Wang, C.M. Saponins of Panax notoginseng: Chemistry, cellular targets and therapeutic opportunities in cardiovascular diseases. Expert Opin. Investig. Drugs 2014, 23, 523–539. [Google Scholar] [CrossRef] [PubMed]
- He, N.W.; Zhao, Y.; Guo, L.; Shang, J.; Yang, X.B. Antioxidant, antiproliferative, and pro-apoptotic activities of a saponin extract derived from the roots of Panax notoginseng (Burk.) F.H. Chen. J. Med. Food. 2012, 15, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Uzayisenga, R.; Ayeka, P.A.; Wang, Y. Anti-diabetic potential of Panax notoginseng saponins (PNS): A review. Phytother. Res. 2014, 28, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Sun, C.H.; Zhao, Y.; Wu, L.J. Hypolipidemic and antioxidant activities of sanchi (radix notoginseng) in rats fed with a high fat diet. Phytomedicine 2011, 18, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Ng, T. Pharmacological activity of sanchi ginseng (Panax notoginseng). J. Pharm. Pharmacol. 2006, 58, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Z.; Anderson, S.; Du, W.; He, T.C.; Yuan, C.S. Red ginseng and cancer treatment. Chin. J. Nat. Med. 2016, 14, 7–16. [Google Scholar] [PubMed]
- Kuzuyama, T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 2002, 66, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Han, J.L.; Li, Z.Q. Molecular cloning, prokaryotic expression and enzyme activity assay of fps from Artemisia annua strain 001. J. Agric. Univ. Hebei 2008, 22, 844–850. [Google Scholar]
- Chen, L.; Lan, X.W.; Li, S.; Zhu, H.; Wu, Y.S. Genetic cloning and sequence analysis of farnesyl pyrophosphate synthase in Panax notoginseng. Chin. Tradit. Herb. Drugs 2006, 37, 1080–1083. [Google Scholar]
- Kim, O.T.; Kim, S.H.; Ohyama, K.; Muranaka, T.; Choi, Y.E.; Lee, H.Y.; Kim, M.Y.; Hwang, B. Upregulation of phytosterol and triterpene biosynthesis in Centella asiatica hairy roots overexpressed ginseng farnesyl diphosphate synthase. Plant Cell Rep. 2010, 29, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, Y.B.; Uddin, M.R.; Lee, S.; Kim, S.U.; Park, S.U. Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase. ACS Synth. Biol. 2014, 3, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhao, S.; Zhang, X. Antisense suppression of cycloartenol synthase results in elevated ginsenoside levels in Panax ginseng hairy roots. Plant Mol. Biol. Rep. 2009, 27, 298–304. [Google Scholar] [CrossRef]
- Helliwell, C.; Waterhouse, P. Constructs and methods for high-throughput gene silencing in plant. Methods 2003, 30, 289–295. [Google Scholar] [CrossRef]
- Wesley, S.V.; Helliwell, C.A.; Smith, N.A.; Wang, M.B.; Rouse, D.T.; Liu, Q.; Gooding, P.S.; Singh, S.P.; Abbott, D.; Stoutjesdijk, P.A.; et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001, 27, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Gan, F.Y.; Zhou, L.G.; Zheng, G.Z.; Wu, K.; Li, L.M. Studies on the callus differentiation from Panax notoginseng. Acta Bot. Yunnan 1993, 15, 61–65. [Google Scholar]
- Zhang, Y.H.; Zhong, J.J.; Yu, J.T. Effects of osmotic pressure on cell growth and production of ginseng saponin and polysaccharide in suspension cultures of Panax notoginseng. Biotechnol. Lett. 1995, 17, 1347–1350. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Zhong, J.J.; Yu, J.T. Effects of nitrogen source on cell growth and production of ginseng saponin and polysaccharide in suspension cultures of Panax notoginseng. Biotechnol. Prog. 1996, 12, 567–571. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, D.Q.; Ge, F.; Yu, G. Effects of co-expression of squalene synthase and dammarenediol-II synthase in Panax notoginseng cells on saponin synthesis. Mod. Food Sci. Technol. 2015, 31, 7–13. [Google Scholar]
- Li, Y.B.; Tang, J.P.; Khatibi, N.H.; Zhu, M.; Chen, D.; Tu, L.; Chen, L.; Wang, S.L. Treatment with ginsenoside Rb1, a component of Panax ginseng, provides neuroprotection in rats subjected to subarachnoid hemorrhage-induced brain injury. Acta Neurochir. Suppl. 2011, 110, 75–79. [Google Scholar] [PubMed]
- Ni, N.; Liu, Q.; Ren, H.; Wu, D.; Luo, C.; Li, P.; Wang, J.B.; Su, H. Ginsenoside Rb1 Protects Rat Neural Progenitor Cells against Oxidative Injury. Molecules 2014, 19, 3012–3024. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Hu, C.J.; Yu, L.Y. Pharmacokinetics of ginsengsides Rg1 and its metabolites in rats. Acta Pharm. Sin. 2010, 45, 636–640. [Google Scholar]
- Ruan, C.C.; Zhang, H.; Zhang, L.X.; Liu, Z.; Sun, G.Z.; Lei, J.; Qin, Y.X.; Zheng, Y.N.; Li, X.; Pan, H.Y. Biotransformation of ginsenoside Rf to Rh1 by recombinant beta-glucosidase. Molecules 2009, 14, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.; Chung, W.S.; Lee, S.K.; Leung, A.W.; Chen, C.H.; Yue, K.K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur. J. Pharmacol. 2006, 550, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Q.; Zhou, P. Advances in studies on ginsenoside Rd. Chin. Tradit. Herb. Drugs 2009, 40, 832–836. [Google Scholar]
- Hofgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.M.; Sun, C.; Sun, Y.Z.; Wu, Q.; Li, Y.; Song, J.Y.; Niu, Y.Y.; Cheng, X.L.; Xu, H.X.; Li, C.Y.; et al. Analysis of the transcriptome of Panax notoginseng root uncover putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genom. 2011, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, Y.S. Initial practice of real time PCR for the expression of SS gene in Panax notoginseng. Guihaia 2008, 28, 703–707. [Google Scholar]
Sample Availability: Samples are not available from the authors. |


| Gene | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| nptII | CTCTGATGCCGCCGTGTT | CCCTGATGCTCTTCGTCCA |
| hptII | GAAGTGCTTGACATTGGGGAAT | AGATGTTGGCGACCTCGTATT |
| Gene | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| FPS | CGGATGCTGGACTATAATGTG | ATTTACGGCAATCATACCAACC |
| CAS | CGGATGGTTTATCTGCCTATGTC | GGTTGCGTGCTTGATTCCA |
| GAPDH | CTACCAACTGTCTTGCTCCCCT | TGATGCAGCTCTTCCACCTCTC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Ge, F.; Sun, Y.; Liu, D.; Chen, C. Strengthening Triterpene Saponins Biosynthesis by Over-Expression of Farnesyl Pyrophosphate Synthase Gene and RNA Interference of Cycloartenol Synthase Gene in Panax notoginseng Cells. Molecules 2017, 22, 581. https://doi.org/10.3390/molecules22040581
Yang Y, Ge F, Sun Y, Liu D, Chen C. Strengthening Triterpene Saponins Biosynthesis by Over-Expression of Farnesyl Pyrophosphate Synthase Gene and RNA Interference of Cycloartenol Synthase Gene in Panax notoginseng Cells. Molecules. 2017; 22(4):581. https://doi.org/10.3390/molecules22040581
Chicago/Turabian StyleYang, Yan, Feng Ge, Ying Sun, Diqiu Liu, and Chaoyin Chen. 2017. "Strengthening Triterpene Saponins Biosynthesis by Over-Expression of Farnesyl Pyrophosphate Synthase Gene and RNA Interference of Cycloartenol Synthase Gene in Panax notoginseng Cells" Molecules 22, no. 4: 581. https://doi.org/10.3390/molecules22040581





