Sampling Terrestrial Environments for Bacterial Polyketides
Abstract
:1. Introduction
2. Unresolved Issues in Microbial Ecology that Limit Bioprospecting for New Polyketides
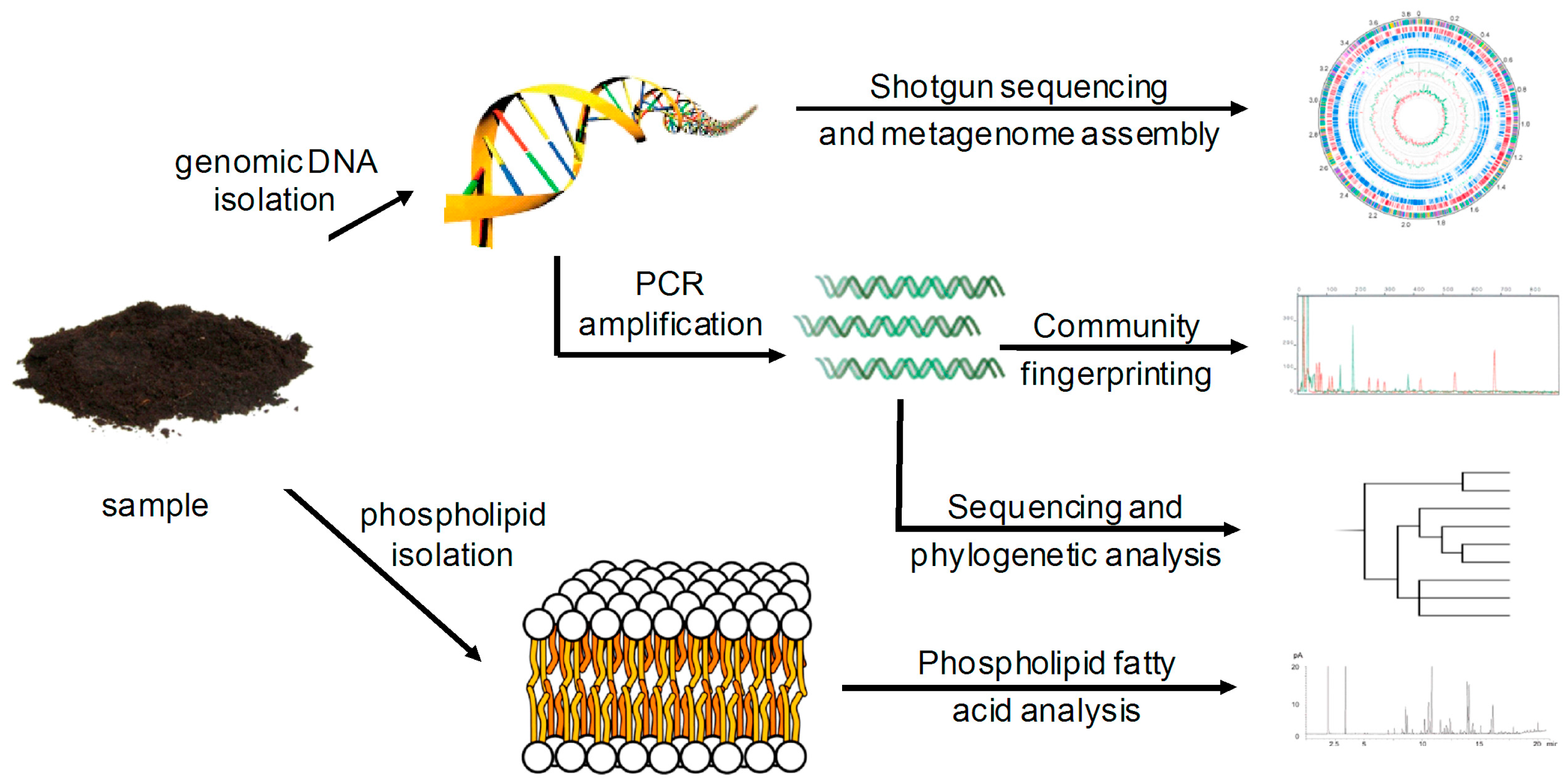
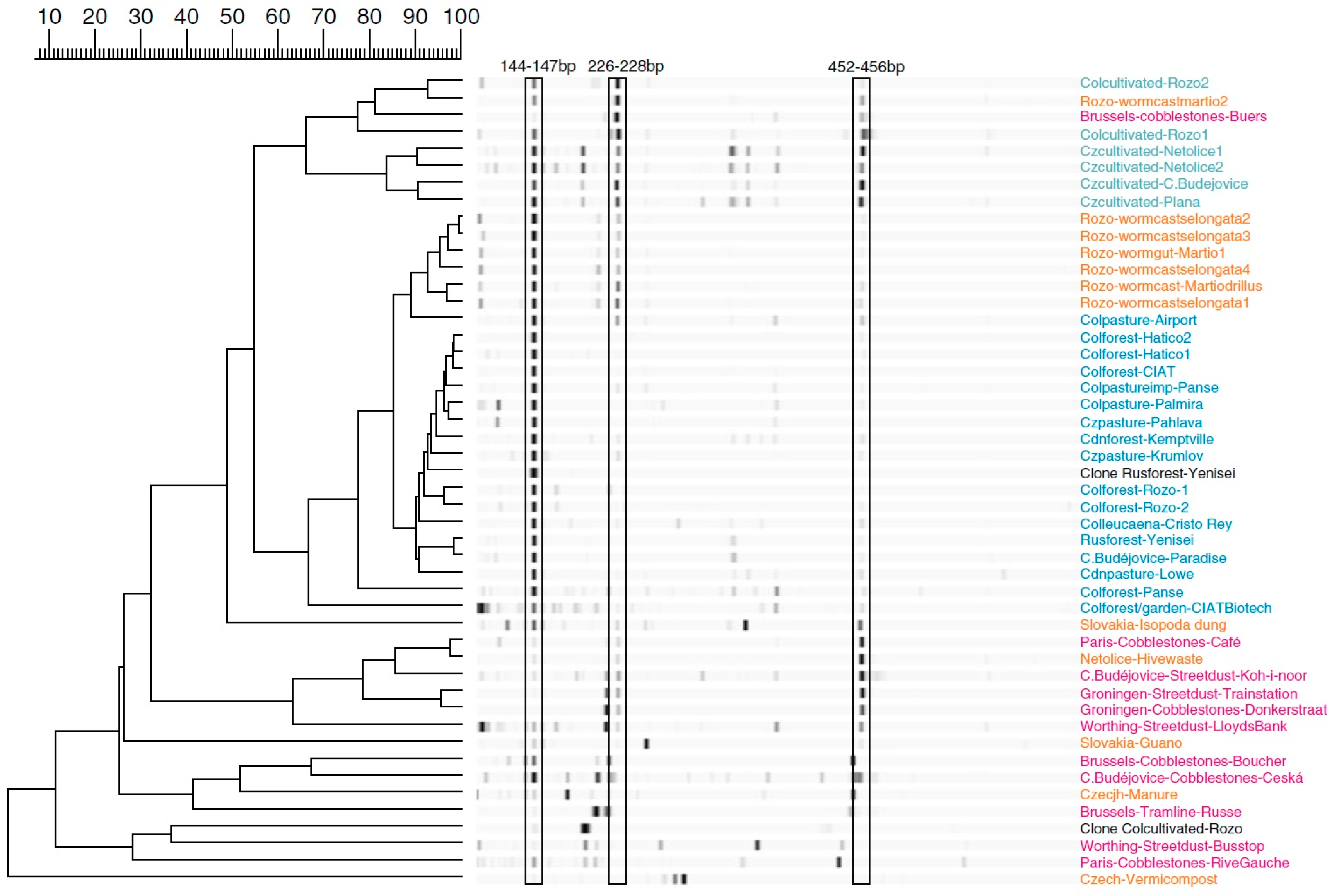
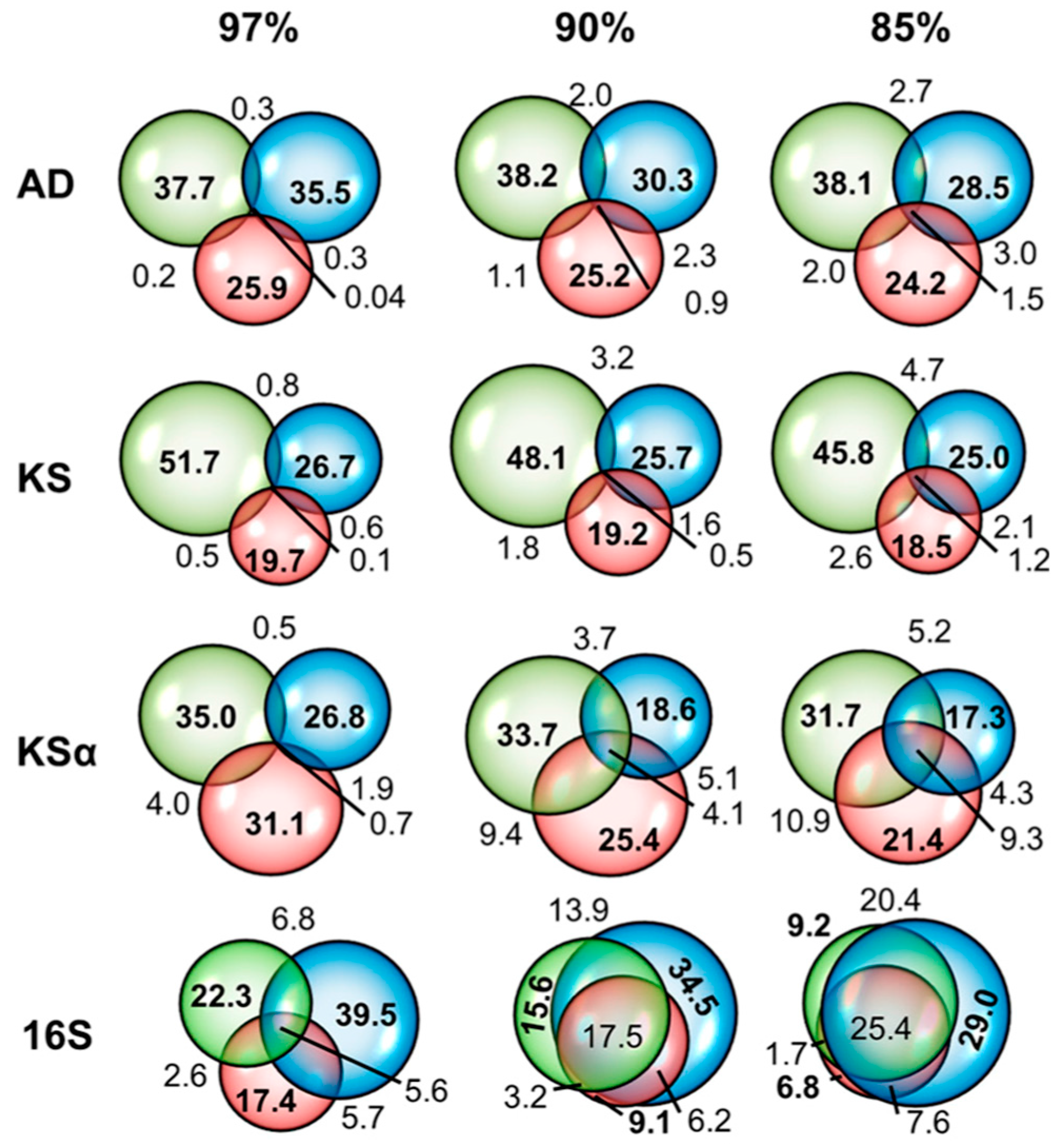
2.1. Data Comparability
2.2. Soil Bacterial Habitats Do Not Correspond to Eukaryotic Habitats
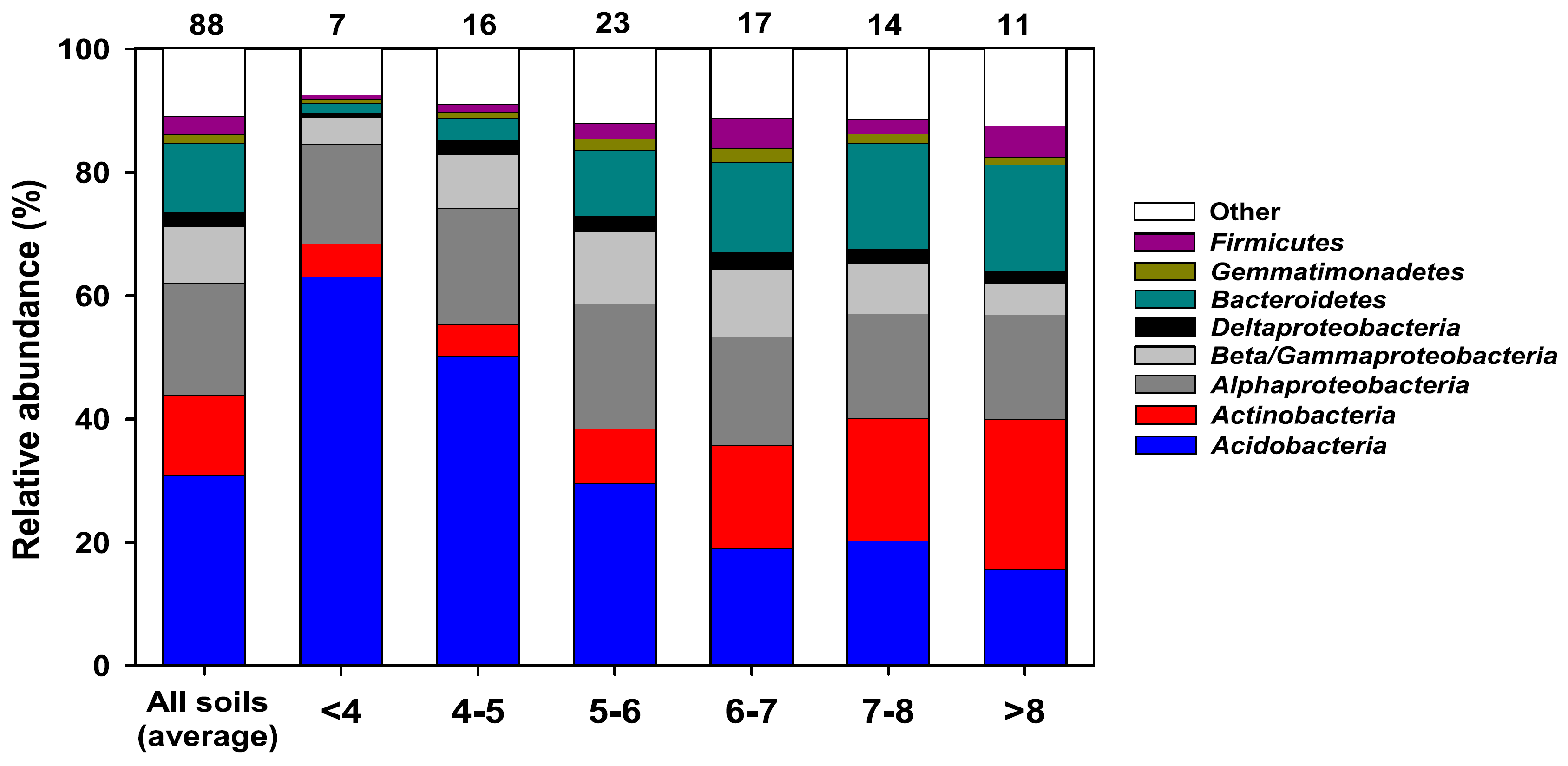
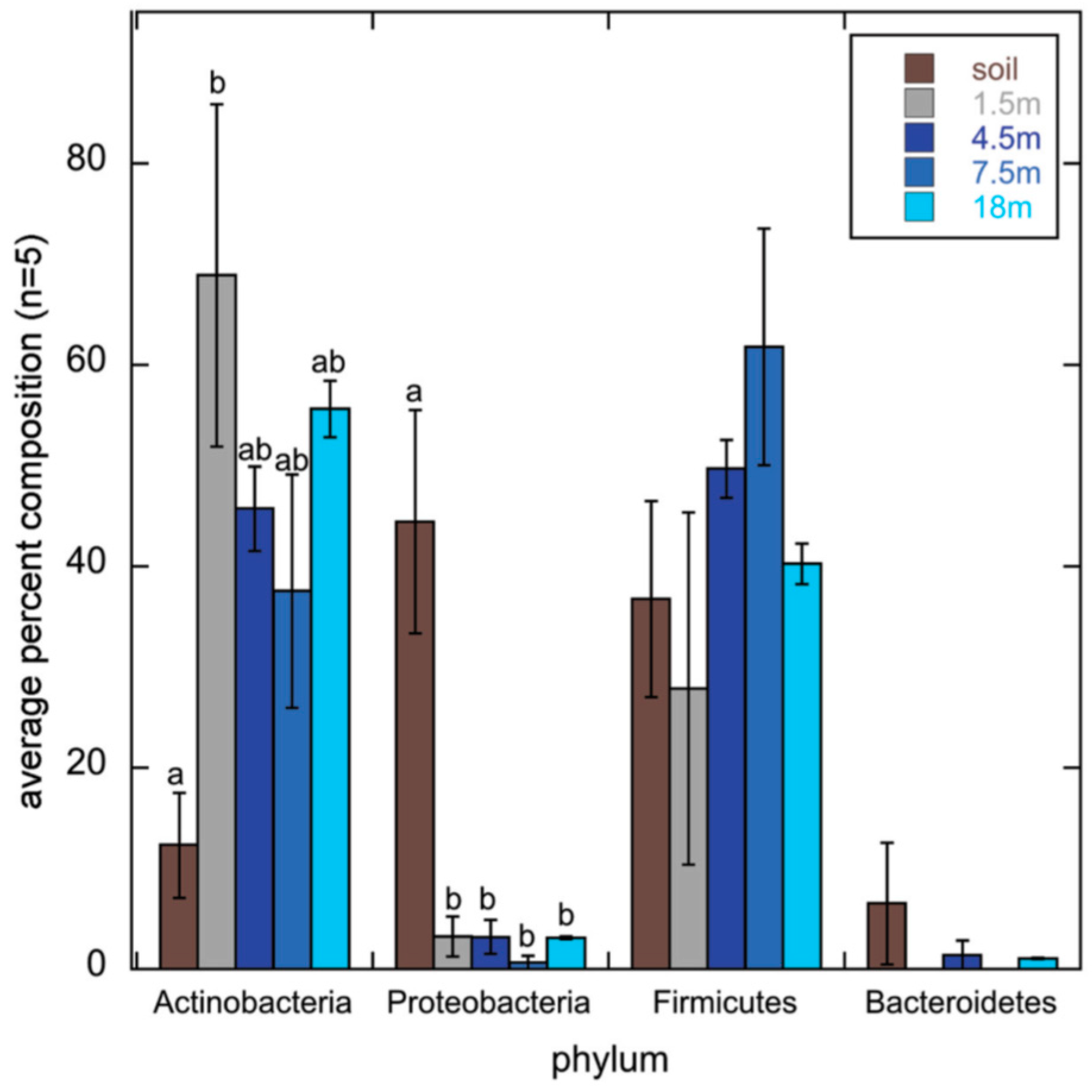
2.3. Actinobacterial Soil Community Structure May Not Follow Bacterial Soil Community Structure
2.4. Non-Actinobacterial Bacterial Polyketide Producers
2.5. The Question of Biogeography
3. Bioprospecting for New Polyketide Discovery
3.1. Terrestrial Environments Where Polyketides Bioprospecting Is Underway
3.1.1. Eukaryotic Associated Bacteria
Insect-Associated Bacteria
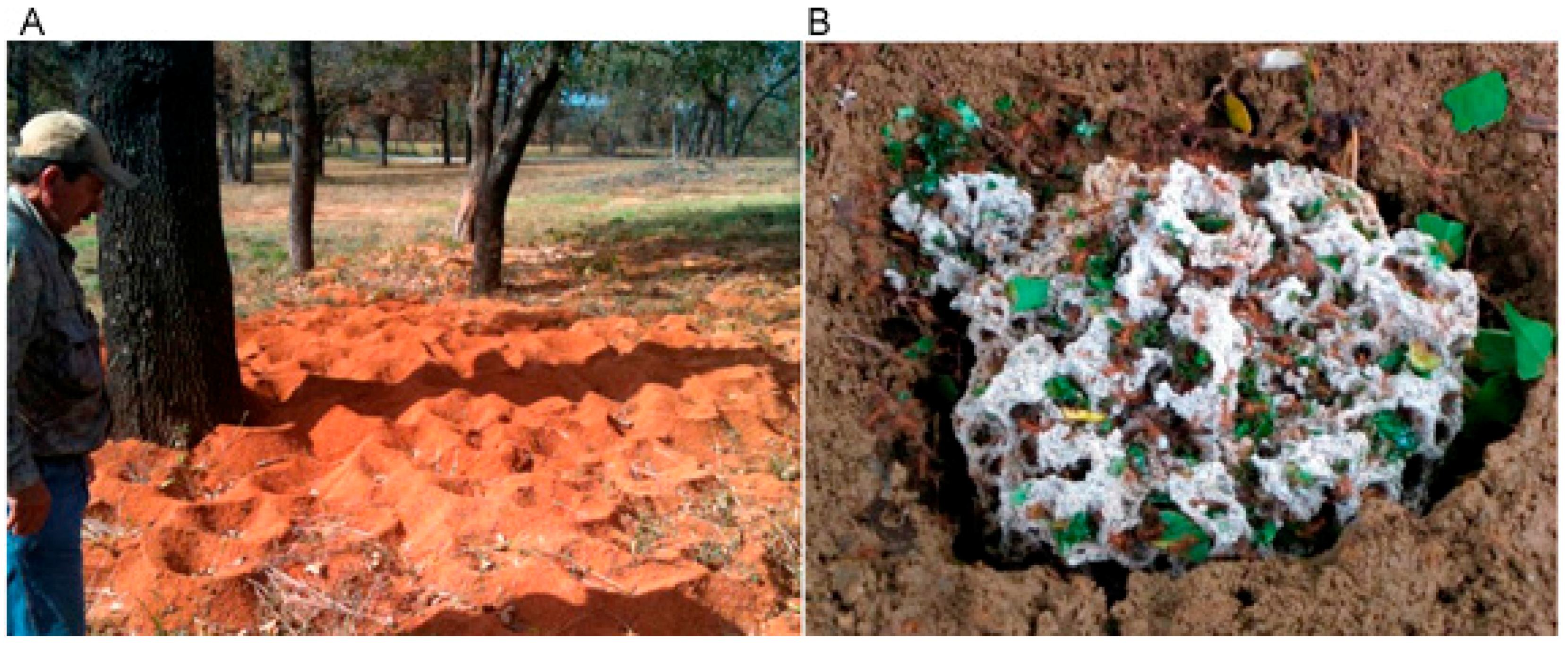
Social Insects as Sources for Bacterial Polyketide Discovery
Non-Social Insects as Sources for Polyketides
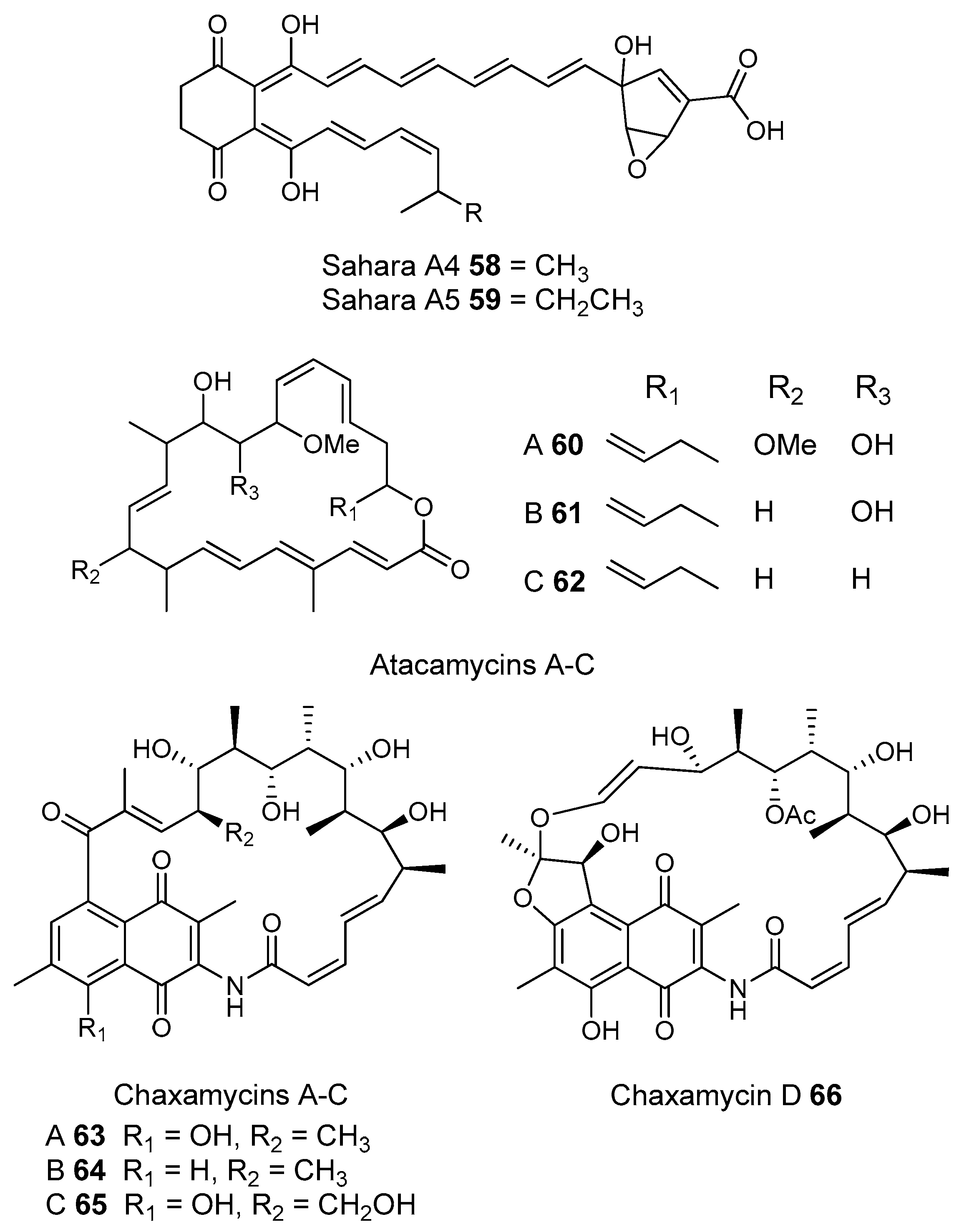
3.1.2. Desert Soils
3.1.3. Disease Suppressive Sediments
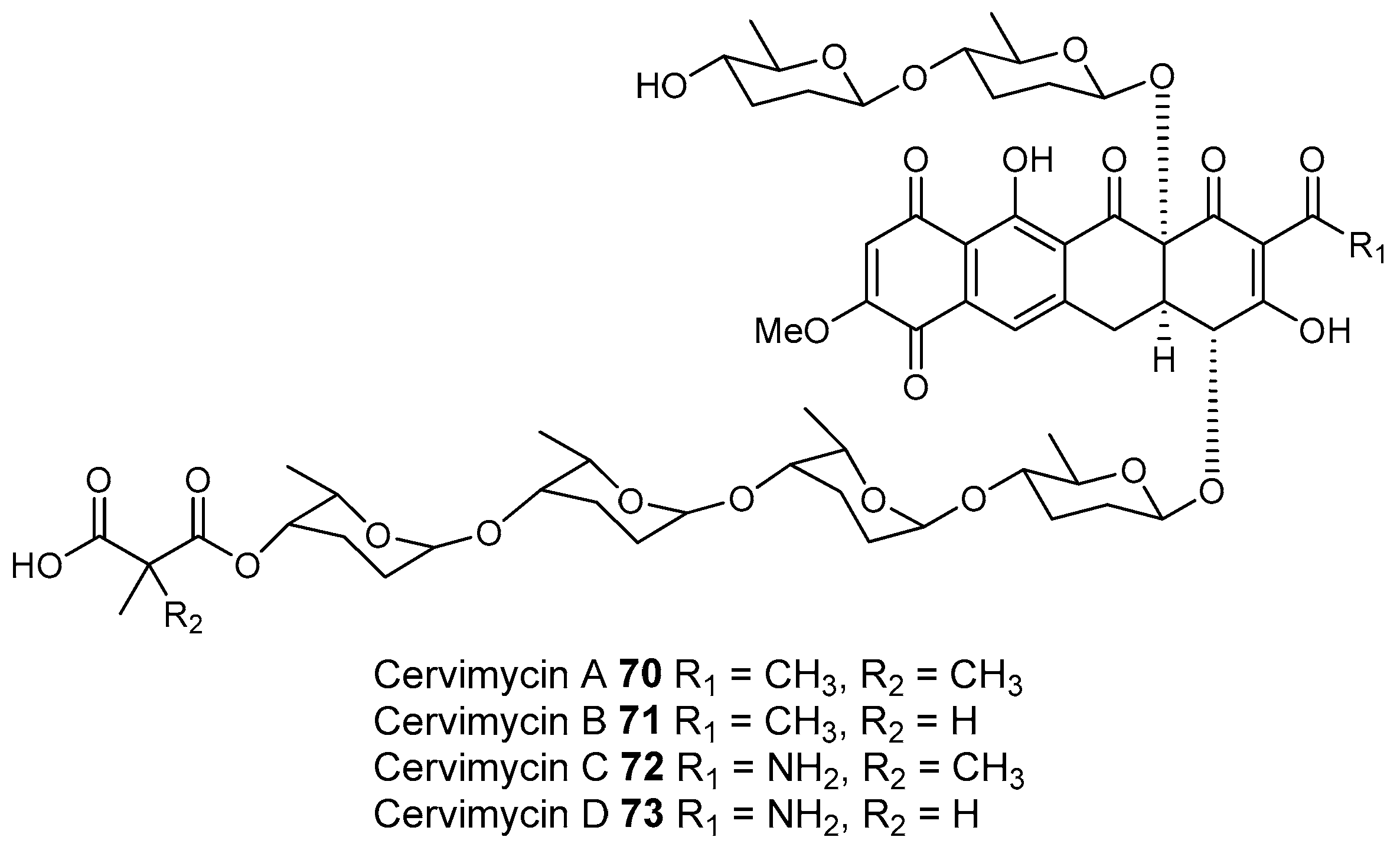
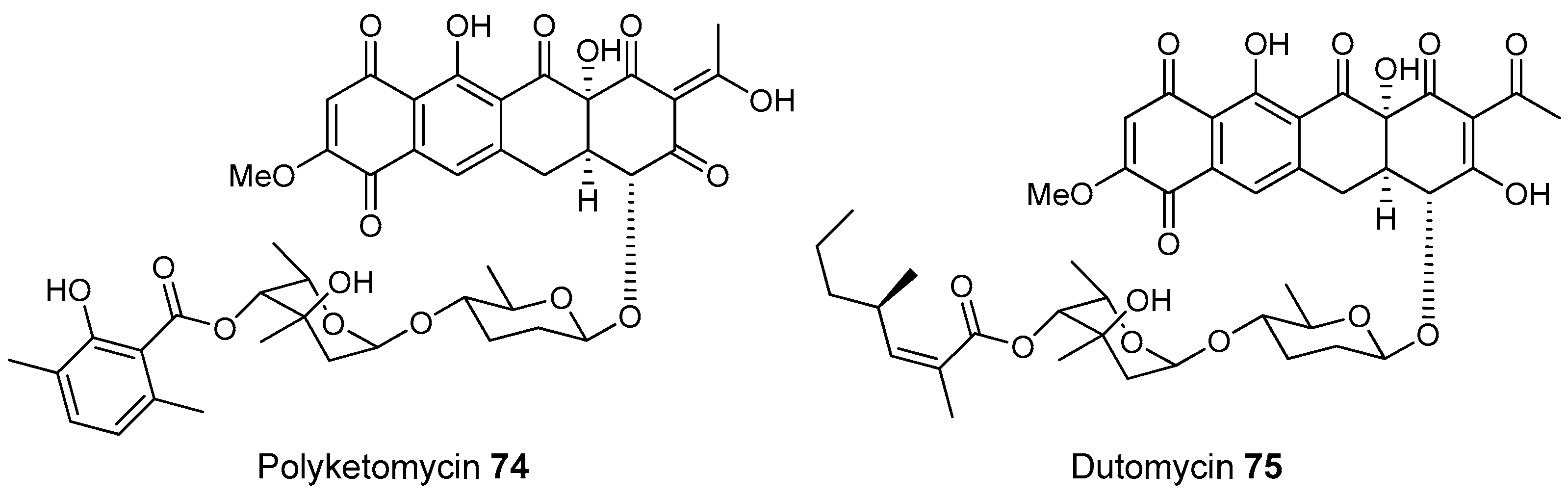
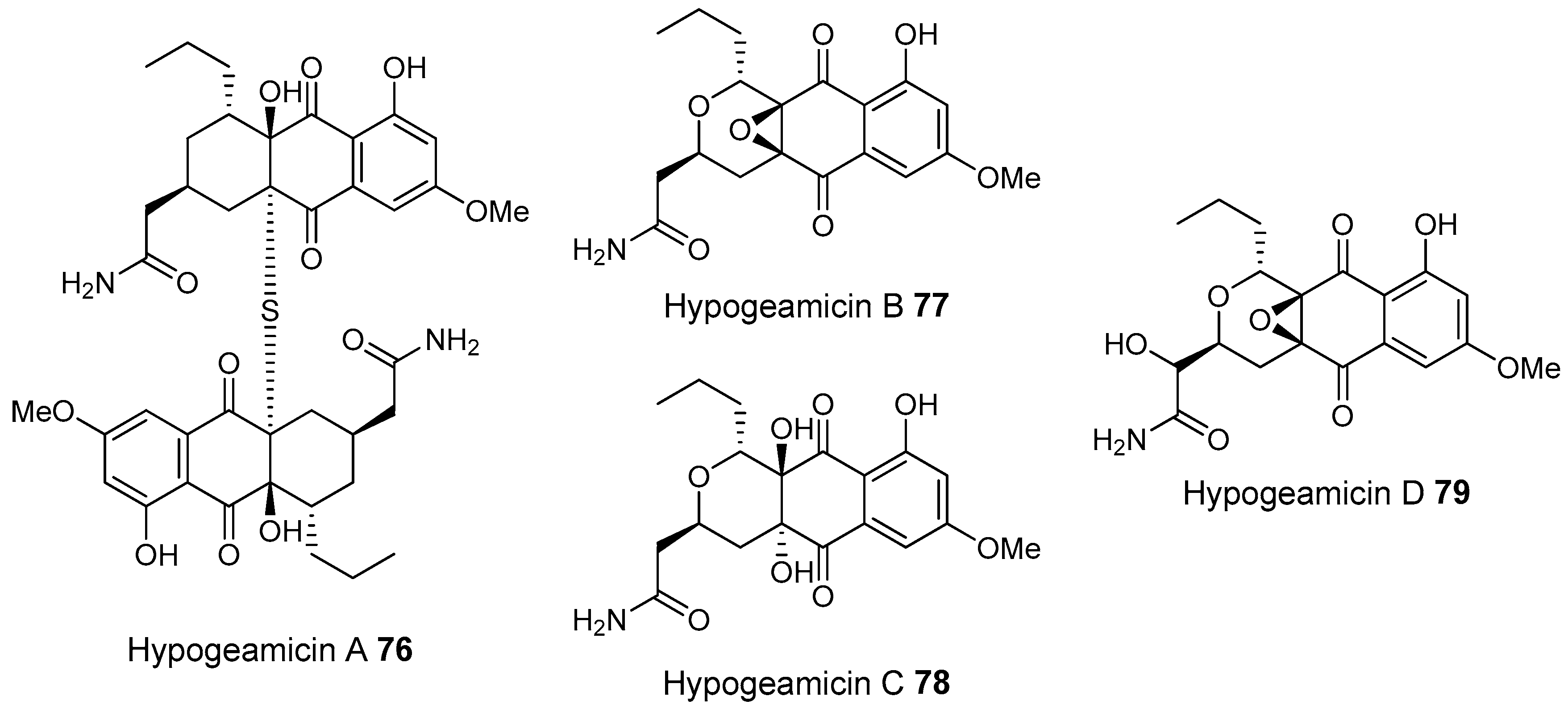
3.1.4. Caves
3.1.5. Extremophiles
3.2. Potential New Environments for Bioprospecting
3.2.1. Cities
3.2.2. Airborne Bacteria
4. Conclusions
4.1. Taking Samples
4.2. Characterising Samples
4.3. Non Actinomycetal Polyketide Producers
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the 30 Years. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, E.J.N.; Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316. [Google Scholar] [CrossRef] [PubMed]
- Horsman, M.E.; Hari, T.P.A.; Boddy, C.N. Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: Logic gate or a victim of fate? Nat. Prod. Rep. 2016, 33, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. Polyketide and nonribosomal peptide antibiotics: Modularity and versatility. Science 2004, 303, 1805–1810. [Google Scholar] [CrossRef] [PubMed]
- Finking, R.; Marahiel, M.A. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 2004, 58, 453–488. [Google Scholar] [CrossRef] [PubMed]
- Felnagle, E.A.; Jackson, E.E.; Chan, Y.A.; Podevels, M.A.; Berti, A.D.; Mcmahon, M.D.; Thomas, M.G.; Podevels, A.M. Nonribosomal Peptide Synthetases Involved in the Production of Medically Relevant Natural Products. Mol. Pharm. 2008, 5, 191–211. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006, 33, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Okada, B.K.; Wu, Y.; Mao, D.; Bushin, L.B.; Seyedsayamdost, M.R. Mapping the Trimethoprim-Induced Secondary Metabolome of Burkholderia thailandensis. ACS Chem. Biol. 2016, 11, 2124–2130. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Lu, C.; Zhang, J.; Zhu, J.; Wang, H.; Shen, Y. Activating a Cryptic Ansamycin Biosynthetic Gene Cluster to Produce Three New Naphthalenic Octaketide Ansamycins with n-Pentyl and n-Butyl Side Chains. Org. Lett. 2015, 17, 3706–3709. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Song, L.; Fox, D.J.; Yeo, V.; Bibb, M.J.; Challis, G.L. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 2012, 3, 2716. [Google Scholar] [CrossRef]
- Stevens, D.C.; Hari, T.P.A.; Boddy, C.N. The role of transcription in heterologous expression of polyketides in bacterial hosts. Nat. Prod. Rep. 2013, 30, 1391–1411. [Google Scholar] [CrossRef] [PubMed]
- Cole Stevens, D.; Henry, M.R.; Murphy, K.A.; Boddy, C.N. Heterologous expression of the oxytetracycline biosynthetic pathway in Myxococcus xanthas. Appl. Environ. Microbiol. 2010, 76, 2681–2683. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Hughes, D.E.; Epstein, S.; Jones, M.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Graziani, E.; Waters, B.; Pan, W.; Li, X.; McDermott, J.; Meurer, G.; Saxena, G.; Andersen, R.J.; Davies, J. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2000, 2, 2401–2404. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Brady, S.F. Arixanthomycins A–C: Phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem. Biol. 2014, 9, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R. Natural Products and the Gene Cluster Revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H. Natural products discovery from micro-organisms in the post-genome era. Biosci. Biotechnol. Biochem. 2017, 81, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. New Approaches to Antimicrobial Discovery. Biochem. Pharmacol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.V.; Ho, J.C.H.; Yadav, G.D.; Yadav, V.G. The Impending Renaissance in Discovery & Development of Natural Products. Curr. Top. Med. Chem. 2017, 17, 251–267. [Google Scholar] [PubMed]
- Zhang, M.M.; Qiao, Y.; Ang, E.L.; Zhao, H. Using natural products for drug discovery: The impact of the genomics era. Expert Opin. Drug Discov. 2017, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, B.; Zhao, H. Breaking the silence: New strategies for discovering novel natural products. Curr. Opin. Biotechnol. 2017, 48, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Methodik, Z.; Lhlung, D.B. Zur methodik der bakterienzählung. Z. Hyg. Infect. 1898, 29, 75–93. [Google Scholar]
- Torsvik, V.; Goksøyr, J.; Daae, F.L.; Torsvik, V.; Goksyr, J.; Daae, F.L. High diversity in DNA of soil bacteria. High Diversity in DNA of Soil Bacteria. Appl. Environ. Microbiol. 1990, 56, 782–787. [Google Scholar] [PubMed]
- Ueda, T.; Suga, Y.; Matsuguchi, M. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur. J. Soil Sci. 1995, 46, 415–421. [Google Scholar] [CrossRef]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial population s by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [PubMed]
- Liu, W.T.; Marsh, T.L.; Cheng, H.; Forney, L.J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997, 63, 4516–4522. [Google Scholar] [PubMed]
- Roesch, L.; Fulthorpe, R.; Riva, A.; Casella, G.; Hadwin, A.; Kent, A.; Daroub, S.; Camargo, F.; Farmerie, W.; Triplett, E. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Tunlid, A.; Hoitink, H.A.; Low, C.; White, D.C. Characterization of bacteria that suppress rhizoctonia damping-off in bark compost media by analysis of Fatty Acid biomarkers. Appl. Environ. Microbiol. 1989, 55, 1368–1374. [Google Scholar] [PubMed]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Kaur, A.; Chaudhary, A.; Kaur, A.; Choudhary, R.; Kaushik, R. Phospholipid fatty acid—A bioindicator of environment monitoring and assessment in soil ecosystem. Curr. Sci. 2005, 89, 1103–1112. [Google Scholar]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Jansson, J.K.; Knight, R. The Earth Microbiome project: Successes and aspirations. BMC Biol. 2014, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010, 12, 2998–3006. [Google Scholar] [CrossRef] [PubMed]
- Parsley, L.C.; Linneman, J.; Goode, A.M.; Becklund, K.; George, I.; Goodman, R.M.; Lopanik, N.B.; Liles, M.R. Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. FEMS Microbiol. Ecol. 2011, 78, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Quaiser, A.; Ochsenreiter, T.; Lanz, C.; Schuster, S.C.; Treusch, A.H.; Eck, J.; Schleper, C. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 2003, 50, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Kim, M.; Singh, D.; Lee-Cruz, L.; Lai-Hoe, A.; Ainuddin, A.N.; Go, R.; Rahim, R.A.; Husni, M.H.A.; Chun, J.; et al. Tropical Soil Bacterial Communities in Malaysia: pH Dominates in the Equatorial Tropics Too. Microb. Ecol. 2012, 64, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.P.; Feng, Y.; Githinji, L.; Ankumah, R.; Balkcom, K.S. Impact of No-tillage and conventional tillage systems on soil microbial communities. Appl. Environ. Soil Sci. 2012, 2012, 548620. [Google Scholar] [CrossRef]
- Neher, D.A.; Weicht, T.R.; Bates, S.T.; Leff, J.W.; Fierer, N. Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PLoS ONE 2013, 8, e79512. [Google Scholar] [CrossRef] [PubMed]
- Delmont, T.O.; Robe, P.; Cecillon, S.; Clark, I.M.; Constancias, F.; Simonet, P.; Hirsch, P.R.; Vogel, T.M. Accessing the soil metagenome for studies of microbial diversity. Appl. Environ. Microbiol. 2011, 77, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Bunge, J.; Leslin, C.; Jeon, S.; Epstein, S.S. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009, 3, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.; Krištůfek, V.; Dijkhuizen, L.; Boddy, C.; Kroetsch, D.; Van Elsas, J.D. Land use intensity controls actinobacterial community structure. Microb. Ecol. 2011, 61, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Bodrossy, L.; Frenzel, P.; Hestnes, A.G.; Krause, S.; Lüke, C.; Meima-Franke, M.; Siljanen, H.; Svenning, M.M.; Bodelier, P.L.E. Impacts of inter- and intralaboratory variations on the reproducibility of microbial community analyses. Appl. Environ. Microbiol. 2010, 76, 7451–7458. [Google Scholar] [CrossRef] [PubMed]
- Cary, S.C.; Fierer, N. The importance of sample archiving in microbial ecology. Nat. Rev. Microbiol. 2014, 12, 789–790. [Google Scholar] [CrossRef] [PubMed]
- Vogel, T.M.; Simonet, P.; Jansson, J.K.; Hirsch, P.R.; Tiedje, J.M.; Elsas, V.; Dirk, J.; Bailey, M.J.; Nalin, R.; Philippot, L. TerraGenome: A consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 2009, 7, 2009. [Google Scholar] [CrossRef]
- Chinese Soil Microbiome Initiative Launches. Available online: http://english.issas.cas.cn/ns/es/201407/t20140702_123686.html (accessed on 24 April 2017).
- Pylro, V.S.; Roesch, L.F.W.; Ortega, J.M.; do Amaral, A.M.; Tótola, M.R.; Hirsch, P.R.; Rosado, A.S.; Góes-Neto, A.; da Costa da Silva, A.L.; Rosa, C.A.; et al. Brazilian Microbiome Project: Revealing the Unexplored Microbial Diversity-Challenges and Prospects. Microb. Ecol. 2014, 67, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Cardon, Z.G.; Cho, M.K.; Dangl, J.L.; Donohue, T.J.; Green, J.L.; Knight, R.; Editor, S.; Maxon, M.E.; Northen, T.R.; et al. Toward a Predictive Understanding of Earth’s Microbiomes to Address 21st Century Challenges. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Campbell, C.D.; Sorenson, S.J.; Zhou, J. Soil genomics. Nat. Rev. Microbiol. 2009, 7, 756. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.I.; Thomson, B.C.; James, P.; Bell, T.; Bailey, M.; Whiteley, A.S. The bacterial biogeography of British soils. Environ. Microbiol. 2011, 13, 1642–1654. [Google Scholar] [CrossRef] [PubMed]
- Dequiedt, S.; Thioulouse, J.; Jolivet, C.; Saby, N.P.A.; Lelievre, M.; Maron, P.A.; Martin, M.P.; Prévost-Bouré, N.C.; Toutain, B.; Arrouays, D.; et al. Biogeographical patterns of soil bacterial communities. Environ. Microbiol. Rep. 2009, 1, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Badger, J.H.; et al. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Birmingham, A.; Knight, R. Context and the human microbiome. Microbiome 2015, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.; Milshteyn, A.; Charlop-Powers, Z.; Brady, S. ESNaPD: A Versatile, Web-Based Bioinformatics Platform for Surveying and Mining Natural Product Biosynthetic Diversity from Metagenomes. Chem. Biol. 2014, 21, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Willig, M.; Kaufman, D.; Stevens, R. Latitudinal gradients of biodiversity: Pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 273–309. [Google Scholar] [CrossRef]
- Fujisaka, S.; Escobar, G.; Veneklaas, E. Plant community diversity relative to human land uses in an Amazon forest colony. Biodivers. Conserv. 1998, 7, 41–57. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, S.; Wardle, D.A.; Lindahl, B.D. Disentangling global soil fungal diversity. Science 2014, 346, 1052–1053. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, A.; Keijzer-Wolters, A.C.; Cacco, G.; van Elsas, J.D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 1999, 38, 1–15. [Google Scholar] [CrossRef]
- Girvan, M.S.; Bullimore, J.; Pretty, J.N.; Mark, A.; Ball, A.S.; Osborn, A.M. Soil Type Is the Primary Determinant of the Composition of the Total and Active Bacterial Communities in Arable Soils Soil Type Is the Primary Determinant of the Composition of the Total and Active Bacterial Communities in Arable Soils. Appl. Environ. Microbiol. 2003, 69, 1800–1809. [Google Scholar] [CrossRef] [PubMed]
- Bossio, D.A.; Girvan, M.S.; Verchot, L.; Bullimore, J.; Borelli, T.; Albrecht, A.; Scow, K.M.; Ball, A.S.; Pretty, J.N.; Osborn, A.M. Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microb. Ecol. 2005, 49, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; de Lima Brossi, M.J.; Kuramae, E.E.; Tsai, S.M. Land-use system shapes soil bacterial communities in Southeastern Amazon region. Appl. Soil Ecol. 2015, 95, 151–160. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Yergeau, E.; Wong, L.C.; Pijl, A.S.; Van Veen, J.A.; Kowalchuk, G.A. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 2012, 79, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, A.; Becker, R. Soil parent material is a key determinant of the bacterial community structure in arable soils. FEMS Microbiol. Ecol. 2006, 56, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lennon, J.T. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 2011, 98, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Dias, R.; de Quadros, P.D.; Davis-Richardson, A.; Camargo, F.A.O.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH Determines Microbial Diversity and Composition in the Park Grass Experiment. Microb. Ecol. 2014, 69, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.N.; Waite, I.S.; Blackburn, A.; Husband, R.; Rushton, S.P.; Manning, D.C.; O’Donnell, A.G. Actinobacterial community dynamics in long term managed grasslands. Antonie Van Leeuwenhoek 2009, 95, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Bartram, A.K.; Jiang, X.; Lynch, M.D.J.; Masella, A.P.; Nicol, G.W.; Dushoff, J.; Neufeld, J.D. Exploring links between pH and bacterial community composition in soils from the Craibstone Experimental Farm. FEMS Microbiol. Ecol. 2014, 87, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Tsai, S.M.; Mendes, L.W.; Faust, K.; De Hollander, M.; Cassman, N.A.; Raes, J.; Van Veen, J.A.; Kuramae, E.E. Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 2015, 24, 2433–2448. [Google Scholar] [CrossRef] [PubMed]
- Montecchia, M.S.; Tosi, M.; Soria, M.A.; Vogrig, J.A.; Sydorenko, O.; Correa, O.S. Pyrosequencing reveals changes in soil bacterial communities after conversion of Yungas forests to agriculture. PLoS ONE 2015, 10, e0119426. [Google Scholar] [CrossRef] [PubMed]
- Waldrop, M.; Balser, T.; Firestone, M. Linking microbial community composition to function in a tropical soil. Soil Biol. Biochem. 2000, 32, 1837–1846. [Google Scholar] [CrossRef]
- Shange, R.S.; Ankumah, R.O.; Ibekwe, A.M.; Zabawa, R.; Dowd, S.E. Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PLoS ONE 2012, 7, e40338. [Google Scholar] [CrossRef] [PubMed]
- Lee-Cruz, L.; Edwards, D.P.; Tripathi, B.M.; Adams, J.M. Impact of logging and forest conversion to oil palm plantations on soil bacterial communities in borneo. Appl. Environ. Microbiol. 2013, 79, 7290–7297. [Google Scholar] [CrossRef] [PubMed]
- Sul, W.J.; Asuming-Brempong, S.; Wang, Q.; Tourlousse, D.M.; Penton, C.R.; Deng, Y.; Rodrigues, J.L.M.; Adiku, S.G.K.; Jones, J.W.; Zhou, J.; et al. Tropical agricultural land management influences on soil microbial communities through its effect on soil organic carbon. Soil Biol. Biochem. 2013, 65, 33–38. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang. Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Lauber, C.L.; Knight, R.; Bradford, M.A.; Fierer, N. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 2010, 91, 3463–3470. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Cassman, N.; de Hollander, M.; Mendes, L.W.; Korevaar, H.; Geerts, R.H.E.M.; van Veen, J.A.; Kuramae, E.E. Impact of long-term N, P, K, and NPK fertilization on the composition and potential functions of the bacterial community in grassland soil. FEMS Microbiol. Ecol. 2014, 90, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Fu, S.; Mathew, R.P. Soil microbial community structure and activity in a 100-year-old fertilization and crop rotation experiment. J. Plant Ecol. 2015, 8, 1–10. [Google Scholar] [CrossRef]
- Warnecke, F.; Amann, R.; Pernthaler, J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 2004, 6, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Humbert, J.F.; Dorigo, U.; Cecchi, P.; Le Berre, B.; Debroas, D.; Bouvy, M. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 2009, 11, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Wawrik, B.; Kerkhof, L.; Zylstra, G.J.; Jerome, J.; Kukor, J.J. Identification of Unique Type II Polyketide Synthase Genes in Soil Identification of Unique Type II Polyketide Synthase Genes in Soil. Appl. Environ. Microbiol. 2005, 71, 2232–2238. [Google Scholar] [CrossRef] [PubMed]
- Wawrik, B.; Kutliev, B.; Abdivasievna, U.A.; Kukor, J.J.; Zylstra, G.J.; Kerkhof, L. Biogeography of actinomycete communities and type II polyketide synthase genes in soils collected in New Jersey and Central Asia. Appl. Environ. Microbiol. 2007, 73, 2982–2989. [Google Scholar] [CrossRef] [PubMed]
- Pang, M.-F.; Tan, G.-Y.A.; Abdullah, N.; Lee, C.-W.; Ng, C.-C. Phylogenetic analysis of type I and type II polyketide synthase from tropical forest soil. Biotechnology 2008, 7, 660–668. [Google Scholar]
- Morlon, H.; O’Connor, T.K.; Bryant, J.A.; Charkoudian, L.K.; Docherty, K.M.; Jones, E.; Kembel, S.W.; Green, J.L.; Bohannan, B.J.M. The biogeography of putative microbial antibiotic production. PLoS ONE 2015, 10, e0130659. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Borsetto, C.; Laskaris, P.; Krsek, M.; Berry, A.E.; Newsham, K.K.; Calvo-Bado, L.; Pearce, D.A.; Vallin, C.; Wellington, E.M.H. Designing and implementing an assay for the detection of rare and divergent NRPS and PKS clones in European, Antarctic and Cuban soils. PLoS ONE 2015, 10, e0138327. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, N.; Zeng, R. Phylogenetic analysis of type I polyketide synthase and nonribosomal peptide synthetase genes in Antarctic sediment. Extremophiles 2008, 12, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, N.; Chen, X.; Jiang, Q.; Zeng, R. Phylogenetic diversity of Type I polyketide synthase genes from sediments of Ardley Island in Antarctica. Acta Oceanol. Sin. 2011, 30, 104–111. [Google Scholar] [CrossRef]
- Zhao, B.; Gao, Z.; Shao, Y.; Yan, J.; Hu, Y.; Yu, J.; Liu, Q.; Chen, F. Diversity analysis of type I ketosynthase in rhizosphere soil of cucumber. J. Basic Microbiol. 2012, 52, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Du, G.-P.; Zhao, Z.-X.; Xie, B.; Li, D.-J. Phylogenetic analysis of type I polyketide synthase and non-ribosomal peptide synthase genes from Mila Mountain in Tibet Plateau. J. Hunan Agric. Univ. 2010, 36, 506–511. [Google Scholar] [CrossRef]
- Ginolhac, A.; Jarrin, C.; Gillet, B.; Robe, P.; Pujic, P.; Tuphile, K.; Bertrand, H.; Vogel, T.M.; Perrière, G.; Simonet, P.; et al. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl. Environ. Microbiol. 2004, 70, 5522–5527. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.S.; Schuch, V.; De Macedo Lemos, E.G. Biotechnology of polyketides: New breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz. J. Microbiol. 2013, 44, 1007–1034. [Google Scholar] [CrossRef] [PubMed]
- Yadav, G.; Gokhale, R.S.; Mohanty, D. SEARCHPKS: A program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003, 31, 3654–3658. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.; Piel, J.; Aris-Brosou, S.; Krištůfek, V.; Boddy, C.N.; Dijkhuizen, L. Habitat-specific type I polyketide synthases in soils and street sediments. J. Ind. Microbiol. Biotechnol. 2014, 41, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Brady, S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. USA 2014, 111, 3757–3762. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.R. The Geographical Distribution of Animals. With a Study of the Relations of Living and Extinct Faunas as Elucidating the Past Changes of the Earth’s Surface; Harper & Brothers: New York, NY, USA, 1876. [Google Scholar]
- Finlay, B.J. Global dispersal of free-living microbial eukaryote species. Science 2002, 296, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- De Wit, R.; Bouvier, T. “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006, 8, 755–758. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, M.A. The nineteenth century roots of “everything is everywhere”. Nat. Rev. Microbiol. 2007, 5, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Bland, J.F.; Esteban, G.F. Ubiquitous Dispersal of Free-Living Microorganisms. In Microbial Diversitiy and Bioprospecting; Bull, A.T., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 216–224. [Google Scholar]
- Martiny, J.B.H.; Bohannan, B.J.M.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R.; et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, R.J.; Grogan, D.W.; Taylor, J.W. Geographic Barriers Isolate Endemic Populations of Hyperthermophilic Archaea. Science 2003, 301, 2002–2004. [Google Scholar] [CrossRef] [PubMed]
- Papke, R.T.; Ramsing, N.B.; Bateson, M.M.; Ward, D.M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 2003, 5, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Tuffin, M.; Cowan, D.A. Biogeography of bacterial communities in hot springs: A focus on the actinobacteria. Extremophiles 2012, 16, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.S. Microbial Diversity of Cave Ecosystems. In Geomicrobiology: Molecular and Environmental Perspective; Barton, L.L., Mandl, M., Loy, A., Eds.; Springer: Dordrecht, The Netherlands, 2010; Chapter 10; pp. 219–238. [Google Scholar]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Zhang, G.; Li, S.; Zhang, W.; Chen, X.; Sun, L.; Zhang, B.; Liu, G.; Chen, T. The diversity and biogeography of the communities of Actinobacteria in the forelands of glaciers at a continental scale. Environ. Res. Lett. 2016, 11, 54012. [Google Scholar] [CrossRef]
- Sun, B.; Wang, F.; Jiang, Y.; Li, Y.; Dong, Z.; Li, Z.; Zhang, X.X. A long-term field experiment of soil transplantation demonstrating the role of contemporary geographic separation in shaping soil microbial community structure. Ecol. Evol. 2014, 4, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.V.B.; Kallifidas, D.; Kim, J.H.; Charlop-Powers, Z.; Feng, Z.; Brady, S.F. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl. Environ. Microbiol. 2012, 78, 3744–3752. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.; Guimaraes, D.O.; De Frias, U.A.; Pupo, M.T.; Seepe, P.; Feng, Z.; Brady, S.F. Global biogeographic sampling of bacterial secondary metabolism. eLife 2015, 2015, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, K.S.; Leff, J.W.; Barberán, A.; Bates, S.T.; Betley, J.; Crowther, T.W.; Kelly, E.F.; Oldfield, E.E.; Shaw, E.A.; Steenbock, C.; et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc. R. Soc. B 2014, 281, 20141988. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Pregitzer, C.C.; Lemetre, C.; Ternei, M.A.; Maniko, J.; Hover, B.M.; Calle, P.Y.; McGuire, K.L.; Garbarino, J.; Forgione, H.M.; et al. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 201615581. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.H.; Thumlert, T.A.; Meisel, L.S.; Carter, P.J.; Kilpin, G.P. “Earthworms downunder”: A survey of the earthworm fauna of urban and agricultural soils in Australia. Soil Biol. Biochem. 1997, 29, 589–597. [Google Scholar] [CrossRef]
- Sterflinger, K.; Prillinger, H. Molecular taxonomy and biodiversity of rock fungal communities in an urban environment (Vienna, Austria). Antonie Van Leeuwenhoek 2001, 80, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Borsetto, C.; Wellington, E.M. Bioprospecting Soil Metagenomes for Antibiotics. In Bioprospecting Success, Potential and Constraints; Paterson, R., Lima, N., Eds.; Springer: Cham, Switzerland, 2017; pp. 113–136. [Google Scholar]
- Gerwick, W.H.; Fenner, A.M. Drug Discovery from Marine Microbes. Microb. Ecol. 2013, 65, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.; Fenical, W. Marine bacterial diversity as a resource for novel microbial products. J. Ind. Microbiol. 1996, 17, 346–351. [Google Scholar] [CrossRef]
- Crawford, J.M.; Clardy, J. Bacterial symbionts and natural products. Chem. Commun. 2011, 47, 7559–7566. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Predominately Uncultured Microbes as Sources of Bioactive Agents. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M. Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 2009, 17, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Aylward, F.O.; Suen, G.; Biedermann, P.H.W.; Adams, A.S.; Scott, J.J.; Malfatti, S.A.; Glavina, T.; Tringe, S.G.; Poulsen, M.; Raffa, K.F.; et al. Convergent Bacterial Microbiotas in the Fungal Agricultural Systems of Insects. MBio 2014, 5, e02077. [Google Scholar] [CrossRef] [PubMed]
- Flórez, L.V.; Biedermann, P.H.W.; Engl, T.; Kaltenpoth, M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 2015, 32, 904–936. [Google Scholar] [CrossRef] [PubMed]
- Menezes, C.; Vollet-Neto, A.; Marsaioli, A.J.; Zampieri, D.; Fontoura, I.C.; Luchessi, A.D.; Imperatriz-Fonseca, V.L. A Brazilian social bee must cultivate fungus to survive. Curr. Biol. 2015, 25, 2851–2855. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Guo, H.; Rischer, M.; Poulsen, M. Natural products from microbes associated with insects. Beilstein J. Org. Chem. 2016, 12, 314–327. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wright, G.D. An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 2011, 22, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Cantley, A.M.; Clardy, J. Animals in a bacterial world: Opportunities for chemical ecology. Nat. Prod. Rep. 2015, 32, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Klassen, J.L. Microbial secondary metabolites and their impacts on insect symbioses. Curr. Opin. Insect Sci. 2014, 4, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, M.P. Access to mutualistic endosymbiotic microbes: An underappreciated benefit of group living. Behav. Ecol. Sociobiol. 2008, 62, 479–497. [Google Scholar] [CrossRef]
- Currie, C.R.; Wong, B.; Stuart, A.E.; Schultz, T.R.; Rehner, S.A.; Mueller, U.G.; Sung, G.-H.; Spatafora, J.W.; Straus, N. A Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 2003, 299, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Currie, C.R.; Scott, J.A.; Summerbell, R.C.; Malloch, D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 1999, 398, 701–704. [Google Scholar] [CrossRef]
- Mueller, U.G.; Dash, D.; Rabeling, C.; Rodrigues, A. Coevolution between attine ants and actinomycete bacteria: A reevaluation. Evolution 2008, 62, 2894–2912. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Ishak, H.D.; Estrada, D.; Dowd, S.E.; Hong, E.; Mueller, U.G. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2009, 106, 17805–17810. [Google Scholar] [CrossRef] [PubMed]
- Caldera, E.J.; Currie, C.R. The Population Structure of Antibiotic-Producing Bacterial Symbionts of Apterostigma dentigerum Ants: Impacts of Coevolution and Multipartite Symbiosis. Am. Nat. 2012, 180, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Van Arnam, E.B.; Ruzzini, A.C.; Sit, C.S.; Horn, H.; Pinto-Tomás, A.A.; Currie, C.R.; Clardy, J. Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc. Natl. Acad. Sci. USA 2016, 113, 12940–12945. [Google Scholar] [CrossRef] [PubMed]
- Kost, C.; Lakatos, T.; Böttcher, I.; Arendholz, W.R.; Redenbach, M.; Wirth, R. Non-specific association between filamentous bacteria and fungus-growing ants. Naturwissenschaften 2007, 94, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.V.; Dillon, R.J.; Dillon, V.M.; Reynolds, S.E.; Samuels, R.I. Ocurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol. Lett. 2004, 239, 319–323. [Google Scholar] [CrossRef] [PubMed]
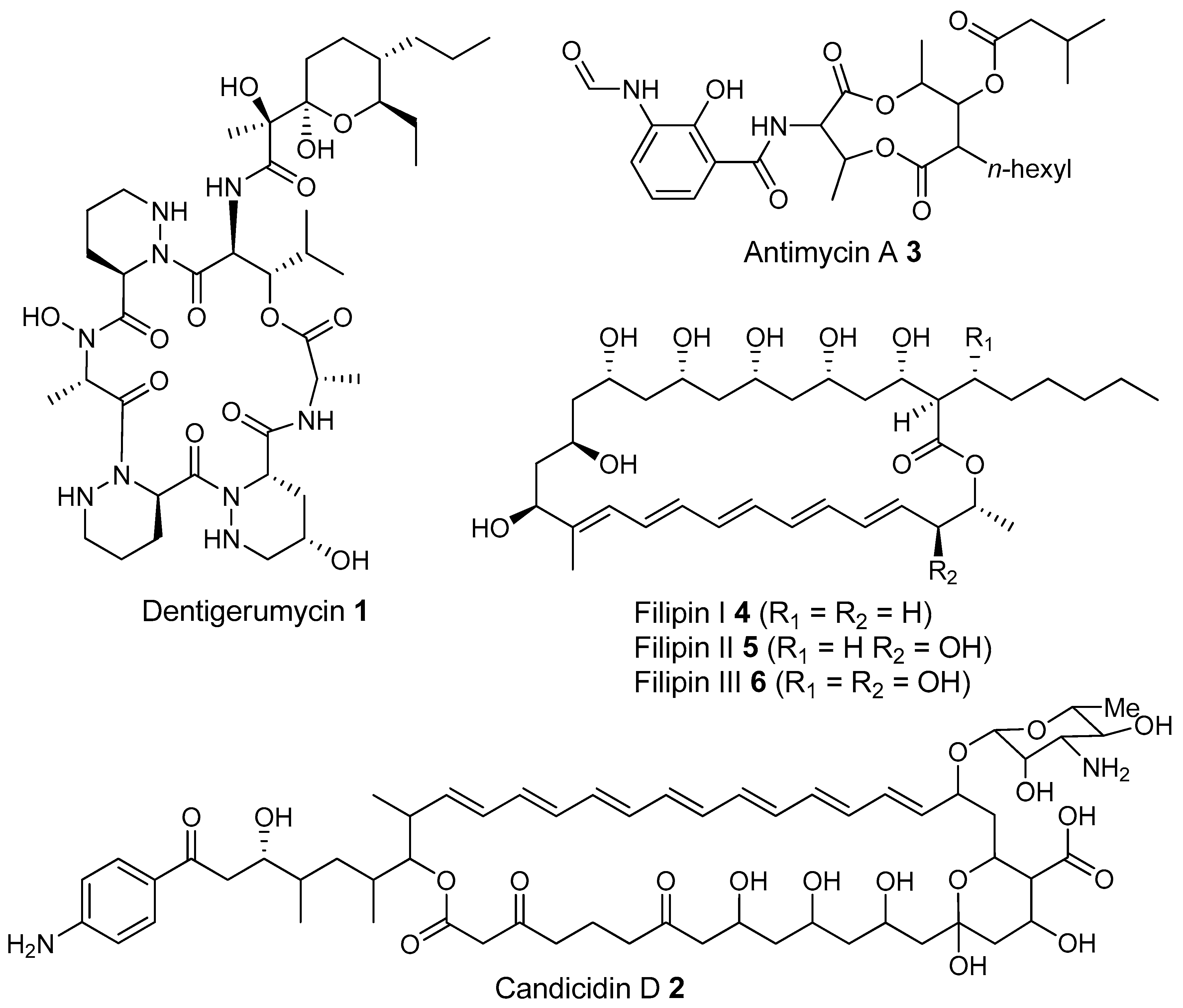
- Oh, D.-C.; Poulsen, M.; Currie, C.R.; Clardy, J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 2009, 5, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Barke, J.; Seipke, R.F.; Grüschow, S.; Heavens, D.; Drou, N.; Bibb, M.J.; Goss, R.J.M.; Yu, D.W.; Hutchings, M.I. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010, 8, 109. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.M.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Grüschow, S.; Barke, J.; Seipke, R.F.; Hill, L.M.; Orivel, J.; Yu, D.W.; Hutchings, M.; Goss, R.J.M. Filipins: The first antifungal “weed killers” identified from bacteria isolated from the trap-ant. RSC Adv. 2014, 4, 57267–57270. [Google Scholar] [CrossRef]
- Aanen, D.K.; Eggleton, P.; Rouland-Lefevre, C.; Guldberg-Froslev, T.; Rosendahl, S.; Boomsma, J.J. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl. Acad. Sci. USA 2002, 99, 14887–14892. [Google Scholar] [CrossRef] [PubMed]
- Visser, A.A.; Nobre, T.; Currie, C.R.; Aanen, D.K.; Poulsen, M. Exploring the Potential for Actinobacteria as Defensive Symbionts in Fungus-Growing Termites. Microb. Ecol. 2012, 63, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Shida, T.; Sasaki, Y.; Kinoshita, N.; Naganawa, H.; Hamada, M.; Takeuchi, T. Vinylamycin, a new depsipeptide antibiotic, from Streptomyces sp. J. Antibiot. 1999, 52, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Carr, G.; Poulsen, M.; Klassen, J.L.; Hou, Y.; Wyche, T.P.; Bugni, T.S.; Currie, C.R.; Clardy, J. Microtermolides A and B from Termite-associated actinomyctes. Org. Lett. 2012, 14, 2822–2825. [Google Scholar] [CrossRef] [PubMed]
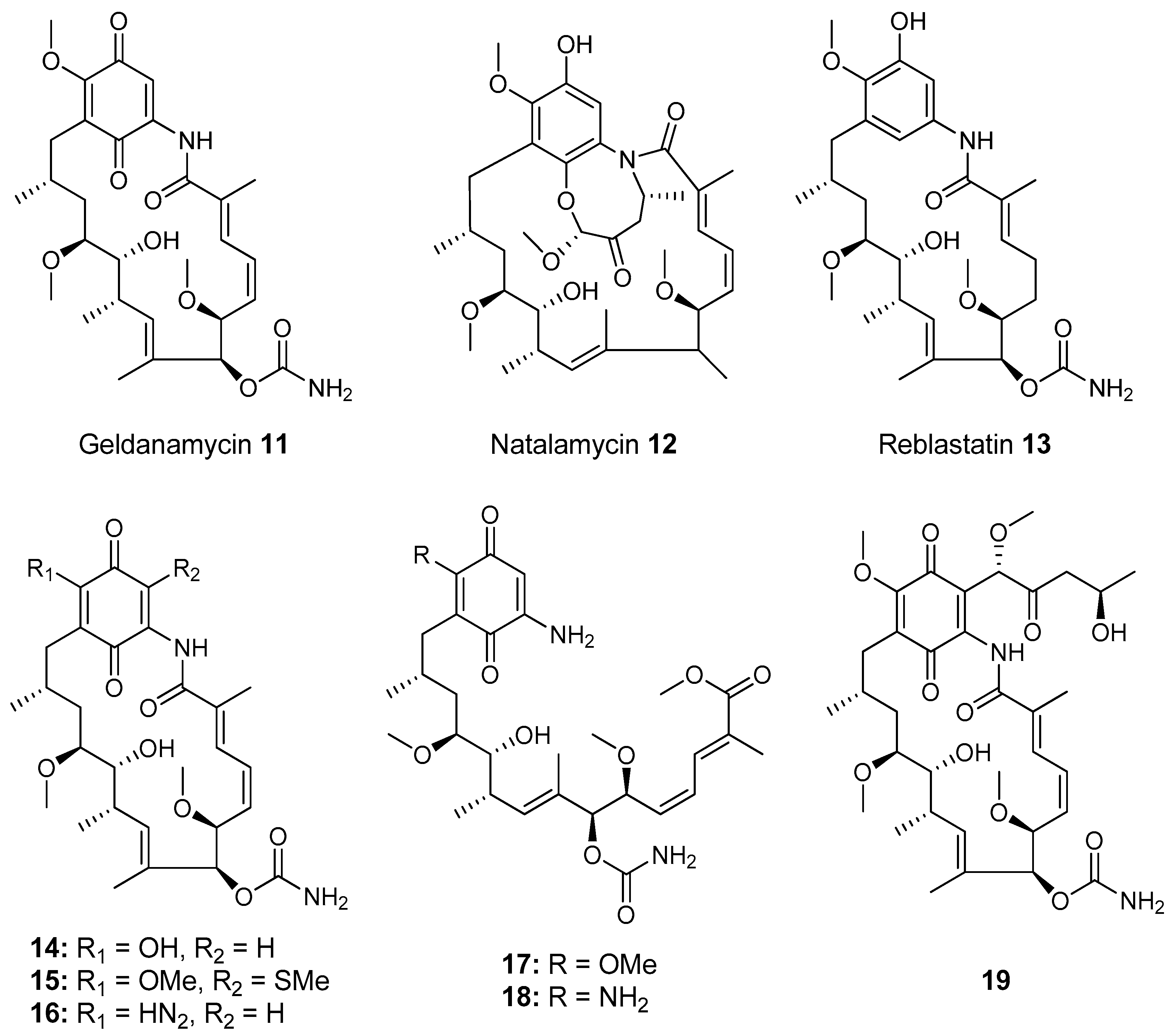
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin A, an Ansamycin from a Termite-Associated Streptomyces sp. Chem. Sci. 2014, 5, 4333–4338. [Google Scholar] [CrossRef] [PubMed]
- Um, S.; Fraimout, A.; Sapountzis, P.; Oh, D.-C.; Poulsen, M. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 2013, 3, 3250. [Google Scholar] [CrossRef] [PubMed]
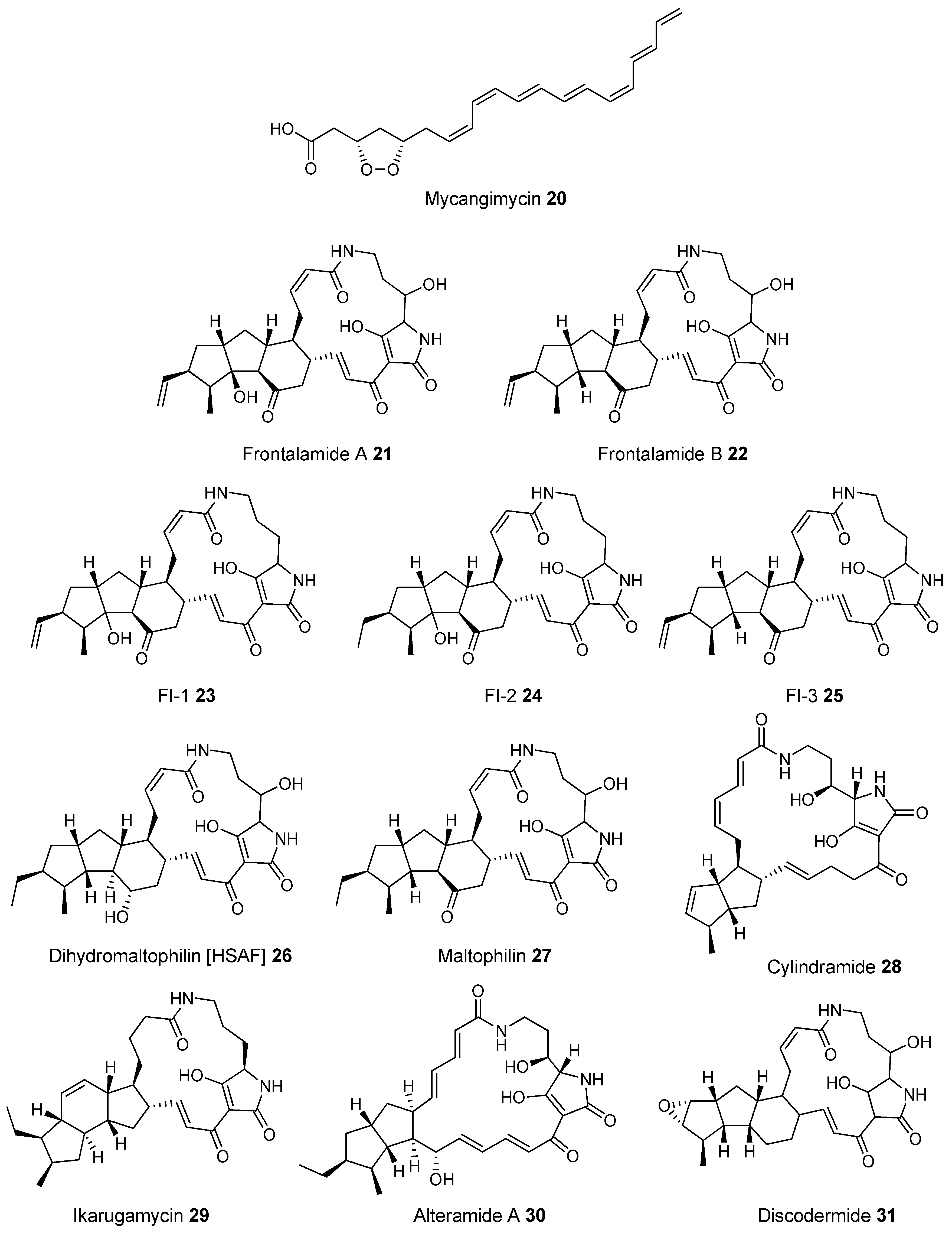
- Scott, J.J.; Oh, D.C.; Yuceer, M.C.; Klepzig, K.D.; Clardy, J. Bacterial Protection of Beetle-Fungus Mutualism. Science 2008, 322, 2008. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zaleta-Rivera, K.; Zhu, X.; Huffman, J.; Millet, J.C.; Harris, S.D.; Yuen, G.; Li, X.C.; Du, L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007, 51, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Blodgett, J.A.; Oh, D.C.; Cao, S.; Currie, C.R.; Kolter, R.; Clardy, J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef] [PubMed]
- Chouvenc, T.; Efstathion, C.A.; Elliott, M.L.; Su, N.-Y. Extended disease resistance emerging from the faecal nest of a subterranean termite. Proc. Biol. Sci. R. Soc. 2013, 280, 20131885. [Google Scholar] [CrossRef] [PubMed]
- Madden, A.A.; Grassetti, A.; Soriano, J.N.; Starks, P.T. Actinomycetes with Antimicrobial Activity Isolated from Paper Wasp (Hymenoptera: Vespidae: Polistinae) Nests. Environ. Entomol. 2013, 42, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Herzner, G.; Engl, T.; Strohm, E. Cryptic combat against competing microbes is a costly component of parental care in a digger wasp. Anim. Behav. 2011, 82, 321–328. [Google Scholar] [CrossRef]
- Kaltenpoth, M.; Gottler, W.; Herzner, G.; Strohm, E. Symbiotic Bacteria Protect Wasp Larvae from Fungal Infestation Martin. Curr. Biol. 2005, 15, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Schmitt, T.; Polidori, C.; Koedam, D.; Strohm, E. Symbiotic streptomycetes in antennal glands of the South American digger wasp genus Trachypus (Hymenoptera, Crabronidae). Physiol. Entomol. 2010, 35, 196–200. [Google Scholar] [CrossRef]
- Kaltenpoth, M.; Yildirim, E.; Gürbüz, M.F.; Herzner, G.; Strohm, E. Refining the roots of the beewolf-streptomyces symbiosis: Antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Appl. Environ. Microbiol. 2012, 78, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Nechitaylo, T.Y.; Westermann, M.; Kaltenpoth, M. Cultivation reveals physiological diversity among defensive “Streptomyces philanthi” symbionts of beewolf digger wasps (Hymenoptera, Crabronidae). BMC Microbiol. 2014, 14, 202. [Google Scholar] [CrossRef] [PubMed]
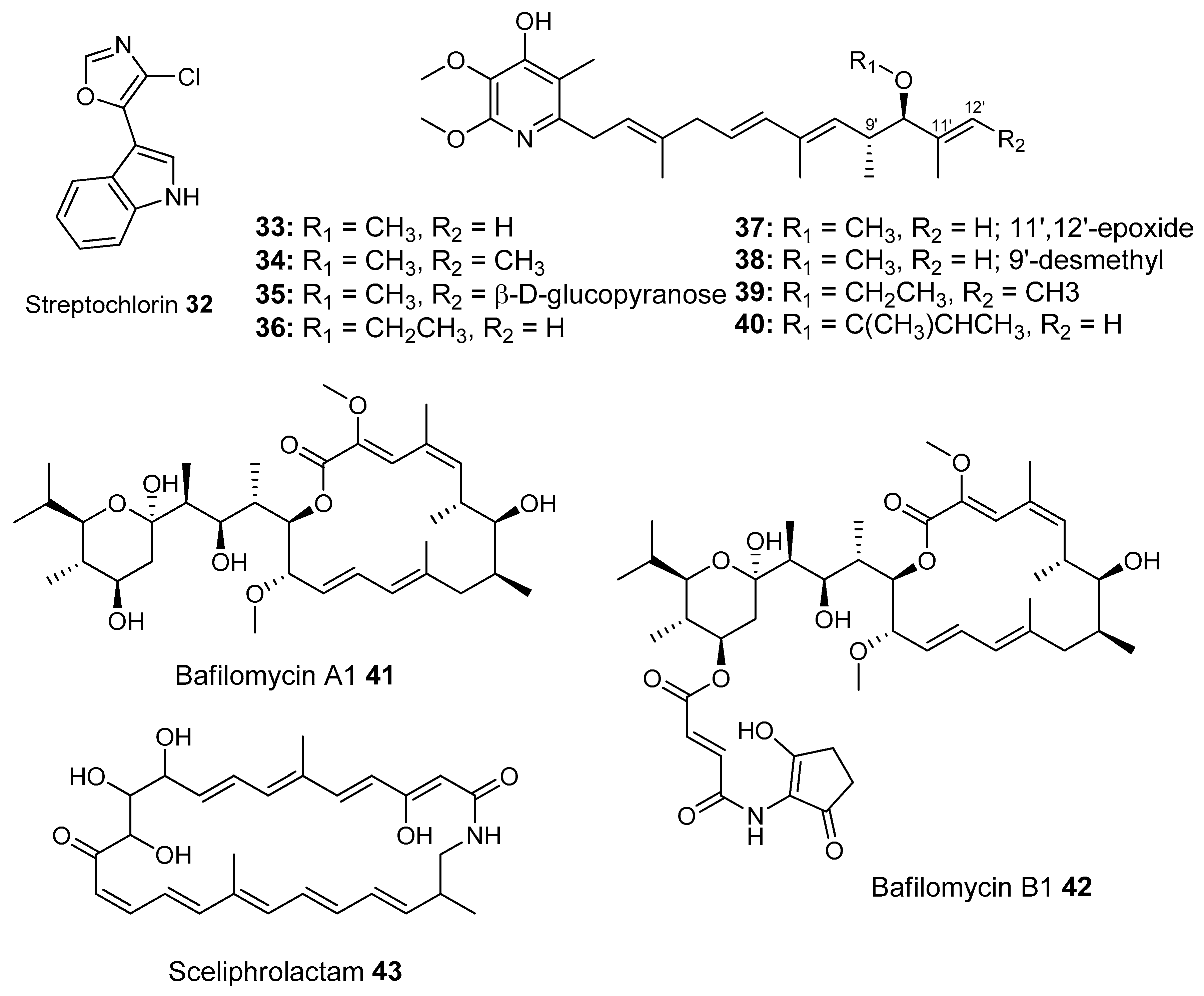
- Shin, H.J.; Jeong, H.S.; Lee, H.S.; Park, S.K.; Kim, H.M.; Kwon, H.J. Isolation and structure determination of streptochlorin, an antiproliferative agent from a marine-derived Streptomyces sp. 04DH110. J. Microbiol. Biotechnol. 2007, 17, 1403–1406. [Google Scholar] [PubMed]
- Takahashi, N.; Suzuki, A.; Tamura, S. Structure of Piericidin A. J. Am. Chem. Soc. 1965, 87, 2066–2068. [Google Scholar] [CrossRef] [PubMed]
- Kroiss, J.; Kaltenpoth, M.; Schneider, B.; Schwinger, M.-G.; Hertweck, C.; Maddula, R.K.; Strohm, E.; Svatos, A. Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 2010, 6, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.; Oh, D.C.; Clardy, J.; Currie, C.R. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 2011, 6, e16763. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.C.; Poulsen, M.; Currie, C.R.; Clardy, J. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org. Lett. 2011, 13, 752–755. [Google Scholar] [CrossRef] [PubMed]
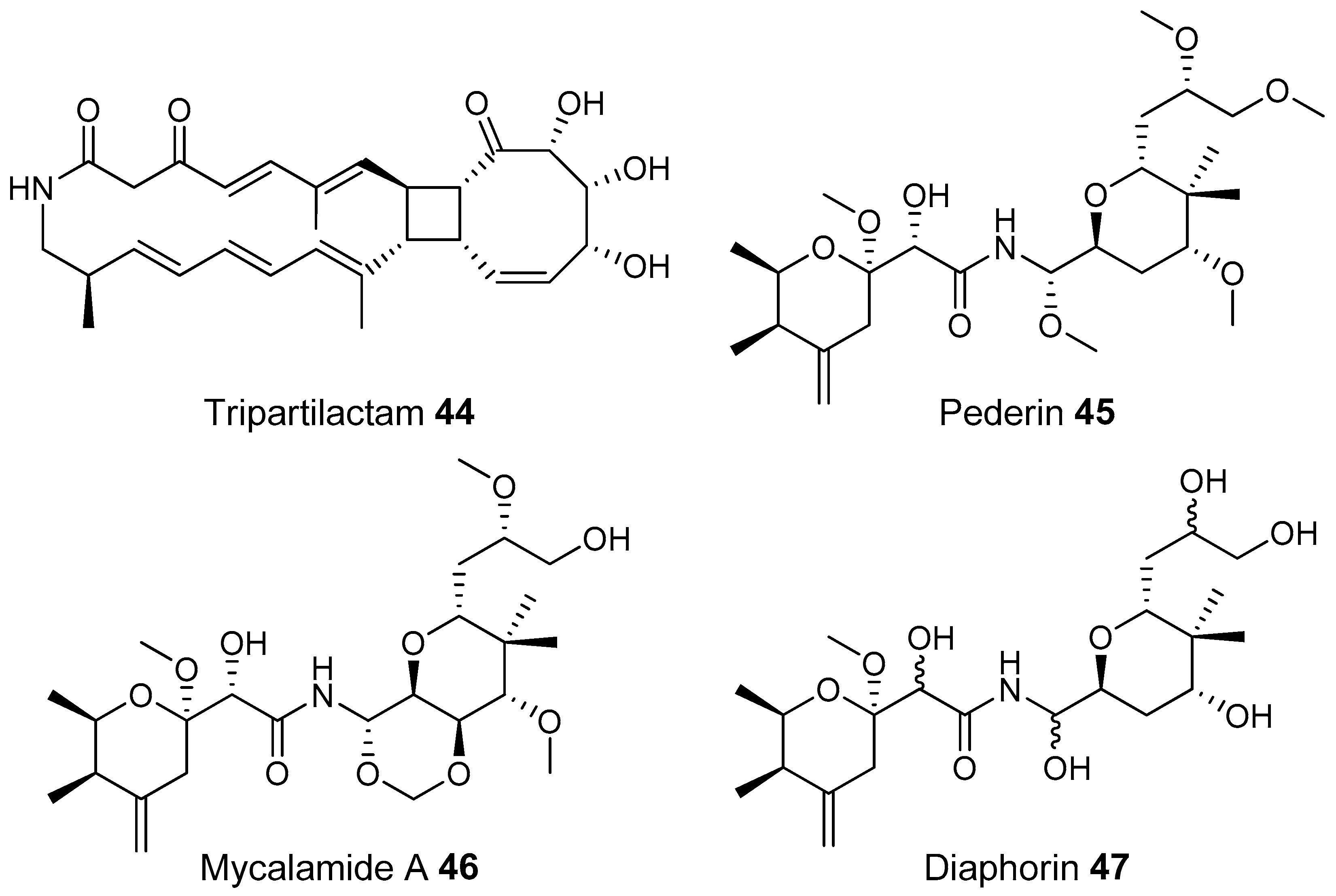
- Park, S.H.; Moon, K.; Bang, H.S.; Kim, S.H.; Kim, D.G.; Oh, K.B.; Shin, J.; Oh, D.C. Tripartilactam, a cyclobutane-bearing tricyclic lactam from a Streptomyces sp. in a dung beetle’s brood ball. Org. Lett. 2012, 14, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Piel, J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 2002, 99, 14002–14007. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; Van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Chapter, I. The Arid Environments. Available online: http://www.fao.org/docrep/t0122e/t0122e03.htm (accessed on 24 April 2017).
- Pointing, S.B.; Chan, Y.; Lacap, D.C.; Lau, M.C.Y.; Jurgens, J.A.; Farrell, R.L. Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl. Acad. Sci. USA 2009, 106, 19964–19969. [Google Scholar] [CrossRef] [PubMed]
- Neilson, J.W.; Quade, J.; Ortiz, M.; Nelson, W.M.; Legatzki, A.; Tian, F.; LaComb, M.; Betancourt, J.L.; Wing, R.A.; Soderlund, C.A.; et al. Life at the hyperarid margin: Novel bacterial diversity in arid soils of the Atacama Desert, Chile. Extremophiles 2012, 16, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Connon, S.A.; Lester, E.D.; Shafaat, H.S.; Obenhuber, D.C.; Ponce, A. Bacterial diversity in hyperarid atacama desert soils. J. Geophys. Res. Biogeosci. 2007, 112, 1–9. [Google Scholar] [CrossRef]
- Chanal, A.; Chapon, V.; Benzerara, K.; Barakat, M.; Christen, R.; Achouak, W.; Barras, F.; Heulin, T. The desert of Tataouine: An extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ. Microbiol. 2006, 8, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Saul-Tcherkas, V.; Steinberger, Y. Soil Microbial Diversity in the Vicinity of a Negev Desert Shrub-Reaumuria negevensis. Microb. Ecol. 2011, 61, 64–81. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial Diversity of Surface Sand Samples from the Gobi and Taklamaken Deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hansen, M.A.; Hansen, L.H.; Jacquiod, S.; Sørensen, S.J. Bioinformatic approaches reveal metagenomic characterization of soil microbial community. PLoS ONE 2014, 9, e93445. [Google Scholar] [CrossRef] [PubMed]
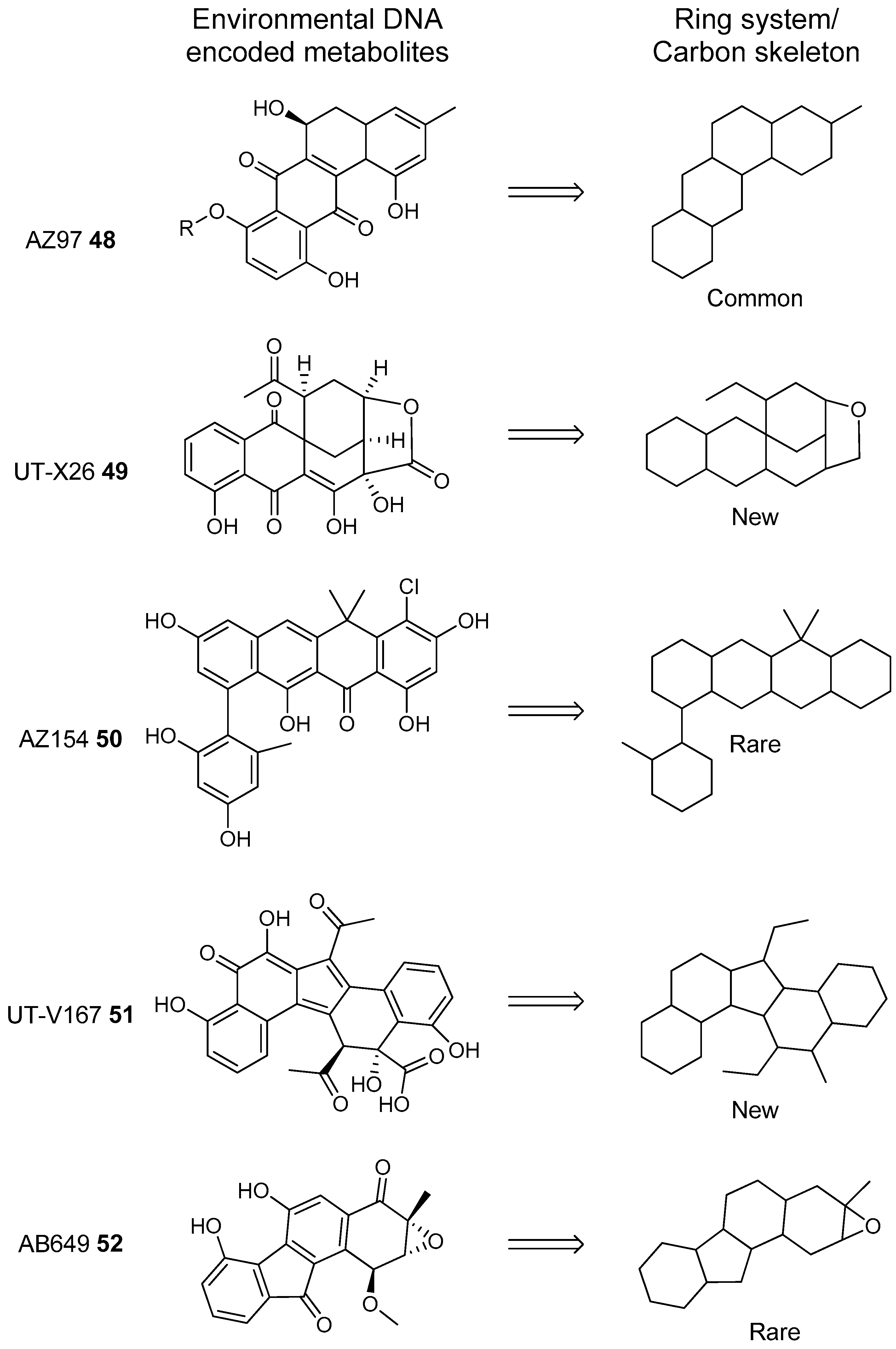
- Owen, J.G.; Reddy, B.V.B.; Ternei, M.A.; Charlop-Powers, Z.; Calle, P.Y.; Kim, J.H.; Brady, S.F. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 2013, 110, 11797–11802. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Kallifidas, D.; Brady, S.F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl. Acad. Sci. USA 2011, 108, 12629–12634. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Kim, J.H.; Brady, S.F. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J. Am. Chem. Soc. 2010, 132, 11902–11903. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.S.; Brady, S.F. Mining soil metagenomes to better understand the evolution of natural product structural diversity: Pentangular polyphenols as a case study. J. Am. Chem. Soc. 2014, 136, 18111–18119. [Google Scholar] [CrossRef] [PubMed]
- Lahoum, A.; Aouiche, A.; Bouras, N.; Verheecke, C.; Klenk, H.-P.; Sabaou, N.; Mathieu, F. Antifungal activity of a Saharan strain of Actinomadura sp. ACD1 against toxigenic fungi and other pathogenic microorganisms. J. Mycol. Méd. 2016, 26, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Boudjella, H.; Bouti, K.; Zitouni, A.; Mathieu, F.; Lebrihi, A.; Sabaou, N. Taxonomy and chemical characterization of antibiotics of Streptosporangium Sg 10 isolated from a Saharan soil. Microbiol. Res. 2006, 161, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Boubetra, D.; Sabaou, N.; Zitouni, A.; Bijani, C.; Lebrihi, A.; Mathieu, F. Taxonomy and chemical characterization of new antibiotics produced by Saccharothrix SA198 isolated from a Saharan soil. Microbiol. Res. 2013, 168, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Silva, B.; Rainey, F.A.; Warren-Rhodes, K.A.; Mckay, C.P.; Navarro-González, R. Atacama Desert Soil Microbiology. In Microbiology of Extreme Soils; Dion, P., Nautiyal, C.S., Eds.; Springer Berlin Heidelberg: Heidelberg, Germany, 2008; Chapter 6; pp. 117–132. [Google Scholar]
- Drees, K.P.; Neilson, J.W.; Betancourt, J.L.; Quade, J.; Henderson, D.A.; Pryor, B.M.; Maier, R.M. Bacterial community structure in the hyperarid core of the Atacama Desert, Chile. Appl. Environ. Microbiol. 2006, 72, 7902–7908. [Google Scholar] [CrossRef] [PubMed]
- Piubeli, F.; de Lourdes Moreno, M.; Kishi, L.T.; Henrique-Silva, F.; García, M.T.; Mellado, E. Phylogenetic Profiling and Diversity of Bacterial Communities in the Death Valley, an Extreme Habitat in the Atacama Desert. Indian J. Microbiol. 2015, 55, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Azua-Bustos, A.; Caro-Lara, L.; Vicuña, R. Discovery and microbial content of the driest site of the hyperarid Atacama Desert, Chile. Environ. Microbiol. Rep. 2015, 7, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, J.; Kulik, A.; Helaly, S.; Bull, A.T.; Goodfellow, M.; Asenjo, J.A.; Maier, A.; Wiese, J.; Imhoff, J.F.; Sussmuth, R.D.; et al. Atacamycins A–C, 22-membered antitumor macrolactones produced by Streptomyces sp. C38*. J. Antibiot 2011, 64, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Houssen, W.E.; Arnold, M.; Abdelrahman, M.H.; Deng, H.; Harrison, W.T.A.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Ferguson, G.; et al. Chaxamycins A–D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J. Nat. Prod. 2011, 74, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Joseph, I.O.F.; Wall, T.E.; Tanner, J.R.; Tawaha, K.; Alali, F.Q.; Li, C.; Oberlies, N.H. Proliferation of antibiotic-producing bacteria and concomitant antibiotic production as the basis for the antibiotic activity of Jordan’s red soils. Appl. Environ. Microbiol. 2009, 75, 2735–2741. [Google Scholar]
- Mazzola, M. Assessment and Management of Soil Microbial Community Structure for Disease Suppression 1. Annu. Rev. Phytopathol. 2004, 42, 35–59. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M. Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie Van Leeuwenhoek 2002, 81, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Janvier, C.; Villeneuve, F.; Alabouvette, C.; Edel-Hermann, V.; Mateille, T.; Steinberg, C. Soil health through soil disease suppression: Which strategy from descriptors to indicators? Soil Biol. Biochem. 2007, 39, 1–23. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Costa, R.; Jansson, J.; Sjöling, S.; Bailey, M.; Nalin, R.; Vogel, T.M.; van Overbeek, L. The metagenomics of disease-suppressive soils—Experiences from the METACONTROL project. Trends Biotechnol. 2008, 26, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Schlatter, D.C.; Kinkel, L.L. Leveraging ecological theory to guide natural product discovery. J. Ind. Microbiol. Biotechnol. 2016, 43, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Adesina, M.F.; Lembke, A.; Costa, R.; Speksnijder, A.; Smalla, K. Screening of bacterial isolates from various European soils for in vitro antagonistic activity towards Rhizoctonia solani and Fusarium oxysporum: Site-dependent composition and diversity revealed. Soil Biol. Biochem. 2007, 39, 2818–2828. [Google Scholar] [CrossRef]
- Garbeva, P.; Postma, J.; Van Veen, J.A.; Van Elsas, J.D. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 2006, 8, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.G.; Otto-Hanson, L.; Lange, A.J.; Bradeen, J.M.; Kinkel, L.L. Plant monocultures produce more antagonistic soil Streptomyces communities than high-diversity plant communities. Soil Biol. Biochem. 2013, 65, 304–312. [Google Scholar] [CrossRef]
- Kinkel, L.L.; Bakker, M.G.; Schlatter, D.C. A coevolutionary framework for managing disease-suppressive soils. Annu. Rev. Phytopathol. 2011, 49, 47–67. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.L.; Lazarovits, G. Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 2003, 72, 169–180. [Google Scholar] [CrossRef]
- Bonanomi, G.; Antignani, V.; Capodilupo, M.; Scala, F. Soil Biology & Biochemistry Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol. Biochem. 2010, 42, 136–144. [Google Scholar]
- Hadar, Y.; Papadopoulou, K.K. Suppressive composts: Microbial ecology links between abiotic environments and healthy plants. Annu. Rev. Phytopathol. 2012, 50, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Boulter, J.I.; Trevors, J.T.; Boland, G.J. Microbial studies of compost: Bacterial identification, and their potential for turfgrass pathogen suppression. World J. Microbiol. Biotechnol. 2002, 18, 661–671. [Google Scholar] [CrossRef]
- Boulter-Bitzer, J.I.; Trevors, J.T.; Boland, G.J. A polyphasic approach for assessing maturity and stability in compost intended for suppression of plant pathogens. Appl. Soil Ecol. 2006, 34, 65–81. [Google Scholar] [CrossRef]
- Bossio, D.A.; Scow, K.M.; Gunapala, N.; Graham, K.J. Determinants of Soil Microbial Communities: Effects of Agricultural Management, Season, and Soil Type on Phospholipid Fatty Acid Profiles. Microb. Ecol. 1998, 36, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Insights perspectives. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.C.G. Compost Tea. In Organic Amendments and Soil Suppressiveness in Plant Disease Management; Meghvansi, M.K., Varma, A., Eds.; Springer International Publishing: Basel, Switzerland, 2015; Chapter 2; pp. 25–49. [Google Scholar]
- Elad, Y.; Shtienberg, D. Effect of compost water extracts on grey mould (Botrytis cinerea). J. Crop Prot. 1994, 13, 109–112. [Google Scholar] [CrossRef]
- Cronin, M.J.; Yohalem, D.S.; Harris, R.F.; Andrew, J.H. Putative Mechanism and Dynamics of Inhibition of the Apple Scab Pathogen Venturia Inaequalis by compost extracts. Soil Biol. Biochem. 1996, 28, 1241–1249. [Google Scholar] [CrossRef]
- Sang, M.K.; Kim, J.; Kim, K.D. Biocontrol Activity and Induction of Systemic Resistance in Pepper by Compost Water Extracts Against Phytophthora capsici. Phytopathology 2010, 100, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Mehta, C.M.; Palni, U.; Franke-Whittle, I.H.; Sharma, A.K. Compost: Its role, mechanism and impact on reducing soil-borne plant diseases. Waste Manag. 2014, 34, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Bradley, G.G.; Punja, Z.K. Composts containing fluorescent pseudomonads suppress fusarium root and stem rot development on greenhouse cucumber. Can. J. Microbiol. 2010, 1995, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, L.S. Identification and Characterization of a Gene Cluster for Synthesis of the Polyketide Antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2–87. J. Bacteriol. 1999, 181, 3155–3163. [Google Scholar]
- Haas, D.; Défago, G. Biological Control of Soil-Borne Pathogens by Fluorescent Pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Iida, T.; Oku, N.; Watanabe, H.; Furihata, K.; Miyanouchi, K. Nomimicin, a new spirotetronate-class polyketide from an actinomycete of the genus Actinomadura. J. Antibiot. 2012, 65, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Meng, J.; Liu, S.; Zhang, H.; Wang, L. Draft Genome Sequence of Streptomyces sp. F-3. Genome Announc. 2013, 4, e00780-16. [Google Scholar]
- Komaki, H.; Ichikawa, N.; Hosoyama, A.; Fujita, N.; Harunari, E.; Igarashi, Y. Draft Genome Sequence of an Anthracimycin Producer, Streptomyces sp. TP-A0875. Genome Announc. 2015, 3, e01149-15. [Google Scholar] [PubMed]
- Bhullar, K.; Waglechner, N.; Pawlowski, A.; Koteva, K.; Banks, E.D.; Johnston, M.D.; Barton, H.A.; Wright, G.D. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 2012, 7, e34953. [Google Scholar] [CrossRef] [PubMed]
- Žák, K.; Světlík, I.V.; Krištůfek, D.; Elhottová, Ľ.; Kováč, A.; Chroňáková, K.; Žák, I. Světlík: The age of bat guano heap in Domica Cave (Slovak Karst NP) and electron microscopy of bat excrements. Acta Carsologica Slovaca 2008, 2, 163–170. [Google Scholar]
- Nováková, A.; Elhottová, D.; Krištůfek, V.; Lukešová, A.; Hill, P.; Kováč, Ľ.; Mock, A.; Ľuptáčik, P. Feeding sources of invertebrates in Ardovská Cave and Domica Cave systems—preliminary results. In Contributions to Soil Zoology in Central Europe I: Proceedings of the 7th Central European Workshop on Soil Zoology; Tajovsky, K., Schlaghamersky, J., Pizl, V., Eds.; Institute of Soil Biology AS CR: Ceske Budejovice, Czech Republic, 2005; pp. 107–112. [Google Scholar]
- Jones, D.S.; Macalady, J.L. The Snotty and the Stringy: Energy for Subsurface Life in Caves. In Their World: A Diversity of Microbial Environments; Hurst, C.J., Ed.; Springer International Publishing: Basel, Switzerland, 2016; Volume 1, Chapter 5; pp. 203–224. [Google Scholar]
- Barton, H.A.; Jurado, V.; Barton, H.A. What’s Up Down There? Microbial Diversity in Caves. ASM Microbe 2007, 2, 132–138. [Google Scholar]
- Jurado, V.; Laiz, L.; Rodriguez-Nava, V.; Boiron, P.; Hermosin, B.; Sanchez-Moral, S.; Saiz-Jimenez, C. Pathogenic and opportunistic microorganisms in caves. Int. J. Speleol. 2010, 39, 15–24. [Google Scholar] [CrossRef]
- Schabereiter-Gurtner, C.; Saiz-Jimenez, C.; Piñar, G.; Lubitz, W.; Rölleke, S. Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonín and La Garma). FEMS Microbiol. Ecol. 2004, 47, 235–247. [Google Scholar] [CrossRef]
- Barton, H.A.; Taylor, N.M.; Kreate, M.P.; Springer, A.C.; Oehrle, S.A.; Bertog, J.L. The impact of host rock geochemistry on bacterial community structure in oligotrophic cave environments. Int. J. Speleol. 2007, 36, 93–104. [Google Scholar] [CrossRef]
- Tomczyk-Żak, K.; Zielenkiewicz, U. Microbial diversity in caves. Geomicrobiol. J. 2016, 33, 20–38. [Google Scholar] [CrossRef]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Groth, I.; Schumann, P.; Laiz, L.; Sanchez-Moral, S.; Cañaveras, J.C.; Saiz-Jimenez, C. Geomicrobiological Study of the Grotta dei Cervi, Porto Badisco, Italy. Geomicrobiol. J. 2001, 18, 241–258. [Google Scholar] [CrossRef]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.D.; Möllmann, U.; Gräfe, U.; Hertweck, C. Cervimycin A–D: A polyketide glycoside complex from a cave bacterium can defeat vancomycin resistance. Chemistry 2005, 11, 5523–5530. [Google Scholar] [CrossRef] [PubMed]
- Herold, K.; Xu, Z.; Gollmick, F.A.; Grafe, U.; Hertweck, C. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Org. Biomol. Chem. 2004, 2, 2411–2414. [Google Scholar] [CrossRef] [PubMed]
- Derewacz, D.K.; McNees, C.R.; Scalmani, G.; Covington, C.L.; Shanmugam, G.; Marnett, L.J.; Polavarapu, P.L.; Bachmann, B.O. Structure and stereochemical determination of hypogeamicins from a cave-derived actinomycete. J. Nat. Prod. 2014, 77, 1759–1763. [Google Scholar] [CrossRef] [PubMed]
- Barton, H.A.; Northup, D.E. Geomicrobiology in cave enviroments: Past, current and future prespectives. J. Cave Karst Stud. 2007, 69, 163–178. [Google Scholar]
- Reinbacher, W.R. Is it gnome, is it berg, is it mont, is it mond? An updated view of the origin and etymology of moonmilk. Bull. Natl. Speleol. Soc. 1994, 56, 1–13. [Google Scholar]
- Axenov-Gibanov, D.V.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Protasov, E.S.; Gamaiunov, S.V.; Rebets, Y.V.; Luzhetskyy, A.N.; Timofeyev, M.A. Actinobacteria Isolated from an Underground Lake and Moonmilk Speleothem from the Biggest Conglomeratic Karstic Cave in Siberia as Sources of Novel Biologically Active Compounds. PLoS ONE 2016, 11, e0149216. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.R.; Dapkevicius, M.L.N.E.; Northup, D.E. Microorganisms isolated from Azorean lava tubes have antimicrobial activity towards food-borne pathogens. In Proceedings of the 9th Food Chemistry Meeting of the Portuguese Society for Chemistry, Angra de Heroismo, Portugal, 29 April 2009; pp. 146–152. [Google Scholar]
- Cheeham, N.; Sadoway, T.; Rule, D.; Watson, K.; Moote, P.; Soliman, L.C.; Azad, N.; Donkor, K.K.; Horne, D. Cure from the cave: Volcanic cave actinomycetes and their potential in drug discovery. Int. J. Speleol. 2013, 42, 35–47. [Google Scholar] [CrossRef]
- Hathaway, J.J.M.; Garcia, M.G.; Balasch, M.M.; Spilde, M.N.; Stone, F.D.; De Lurdes, M.; Dapkevicius, N.E.; Amorim, D.E.I.R.; Gabriel, R.; Borges, P.A.V.; et al. Comparison of Bacterial Diversity in Azorean and Hawai’ian Lava Cave Microbial Mats. Geomicrobiol. J. 2014, 313, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, C.; Marshall Hathaway, J.J.; Enes Dapkevicius, M.d.L.N.; Miller, A.Z.; Kooser, A.; Northup, D.E.; Jurado, V.; Fernandez, O.; Saiz-Jimenez, C.; Cheeptham, N. Actinobacterial Diversity in Volcanic Caves and Associated Geomicrobiological Interactions. Front. Microbiol. 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Robinson, S.; Jebakumar, D. Unexplored hypersaline habitats are sources of novel actinomycetes. Front. Microbiol 2014, 5, 242. [Google Scholar] [CrossRef] [PubMed]
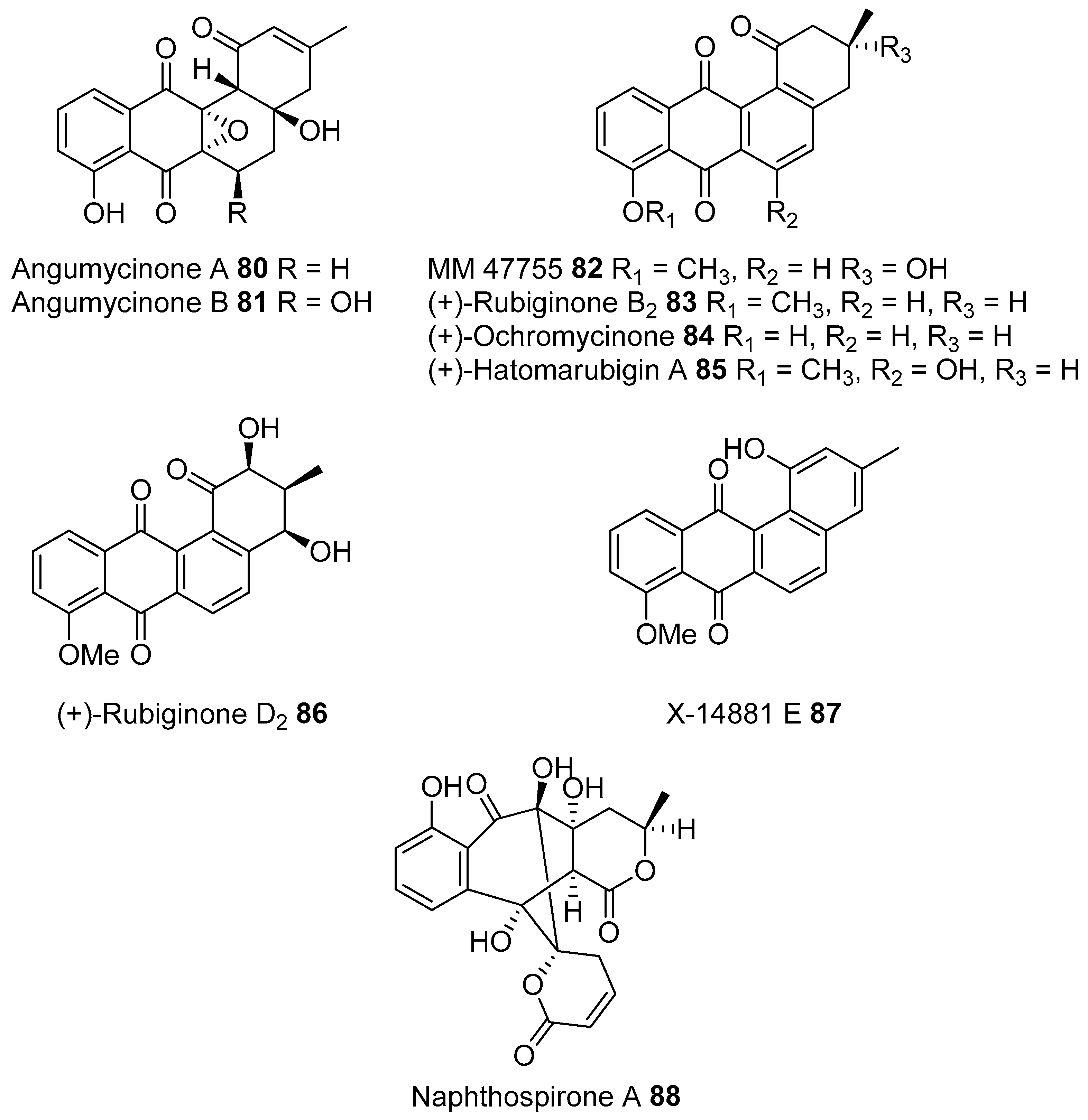
- Park, H.B.; Lee, J.K.; Lee, K.R.; Kwon, H.C. Angumycinones A and B, two new angucyclic quinones from Streptomyces sp. KMC004 isolated from acidic mine drainage. Tetrahedron Lett. 2014, 55, 63–66. [Google Scholar] [CrossRef]
- Ding, Z.G.; Li, M.G.; Zhao, J.Y.; Ren, J.; Huang, R.; Xie, M.J.; Cui, X.L.; Zhu, H.J.; Wen, M.L. Naphthospironone A: An unprecedented and highly functionalized polycyclic metabolite from an alkaline mine waste extremophile. Chemistry 2010, 16, 3902–3905. [Google Scholar] [CrossRef] [PubMed]
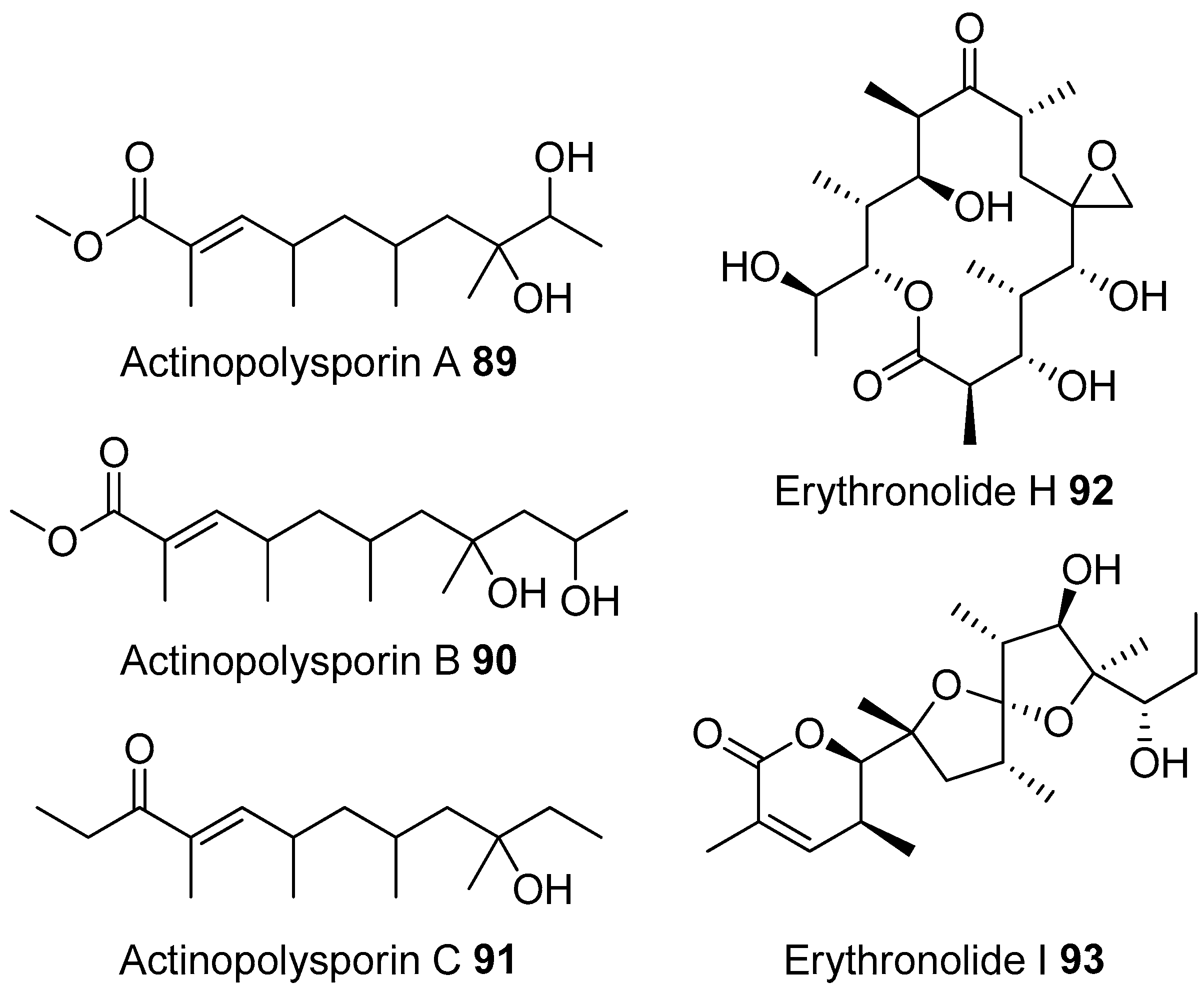
- Tang, S.K.; Wang, Y.; Klenk, H.P.; Shi, R.; Lou, K.; Zhang, Y.J.; Chen, C.; Ruan, J.S.; Li, W.J. Actinopolyspora alba sp. nov. and Actinopolyspora erythraea sp. nov., isolated from a salt field, and reclassification of Actinopolyspora iraqiensis Ruan et al. 1994 as a heterotypic synonym of Saccharomonospora halophila. Int. J. Syst. Evol. Microbiol. 2011, 61, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.X.; Huang, S.X.; Tang, S.K.; Jiang, C.L.; Duan, Y.; Beutler, J.A.; Henrich, C.J.; McMahon, J.B.; Schmid, T.; Blees, J.S.; et al. Actinopolysporins A–C and tubercidin as a pdcd4 stabilizer from the halophilic actinomycete Actinopolyspora erythraea YIM 90600. J. Nat. Prod. 2011, 74, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- McKinney, M.L. Effects of urbanization on species richness: A review of plants and animals. Urban Ecosyst. 2008, 11, 161–176. [Google Scholar] [CrossRef]
- Faeth, S.H.; Bang, C.; Saari, S. Urban biodiversity: Patterns and mechanisms. Ann. N. Y. Acad. Sci. 2011, 1223, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Erséus, C.; Grimm, R.; Healy, B.; Lundberg, S.; Rota, E.; Timm, T. Clitellate diversity in Nationalstadsparken, an urban national park in Stockholm, Sweden. Hydrobiologia 1999, 406, 101–110. [Google Scholar] [CrossRef]
- Fang, Y.; Yoh, M.; Koba, K.; Zhu, W.; Takebayashi, Y.; Xiao, Y.; Lei, C.; Mo, J.; Zhang, W.; Lu, X. Nitrogen deposition and forest nitrogen cycling along an urban-rural transect in southern China. Glob. Chang. Biol. 2011, 17, 872–885. [Google Scholar] [CrossRef]
- McDonnell, M.J.; Pickett, S.T.A.; Groffman, P.; Bohlen, P.; Pouyat, R.V.; Zipperer, W.C.; Parmelee, R.W.; Carreiro, M.M.; Medley, K. Ecosystem processes along an urban to rural gradient. Urban Ecosyst. 1997, 1, 21–36. [Google Scholar] [CrossRef]
- Gosse, J.T.; Hill, P.; Dowd, S.E.; Boddy, C.N. Draft Genome Sequence of Streptomyces sp. Strain PBH53, Isolated from an Urban Environment. Genome Announc. 2015, 3, e00859-15. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.F.; Werth, J.T. Is the lower atmosphere a readily accessible reservoir of culturable, antimicrobial compound-producing Actinomycetales? Front. Microbiol. 2015, 6, 802. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.I.; Thomson, B.C.; Plassart, P.; Gweon, H.S.; Stone, D.; Creamer, R.E.; Lemanceau, P.; Bailey, M.J. Mapping and validating predictions of soil bacterial biodiversity using European and national scale datasets. Appl. Soil Ecol. 2016, 97, 61–68. [Google Scholar] [CrossRef]
- Levy, S.E.; Myers, R.M. Advancements in Next-Generation Sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M.H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [PubMed]
- Monciardini, P.; Sosio, M.; Cavaletti, L.; Chiocchini, C.; Stefano, D. New PCR primers for the selective amplication of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 2002, 42, 419–429. [Google Scholar] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hill, P.; Heberlig, G.W.; Boddy, C.N. Sampling Terrestrial Environments for Bacterial Polyketides. Molecules 2017, 22, 707. https://doi.org/10.3390/molecules22050707
Hill P, Heberlig GW, Boddy CN. Sampling Terrestrial Environments for Bacterial Polyketides. Molecules. 2017; 22(5):707. https://doi.org/10.3390/molecules22050707
Chicago/Turabian StyleHill, Patrick, Graham W. Heberlig, and Christopher N. Boddy. 2017. "Sampling Terrestrial Environments for Bacterial Polyketides" Molecules 22, no. 5: 707. https://doi.org/10.3390/molecules22050707
APA StyleHill, P., Heberlig, G. W., & Boddy, C. N. (2017). Sampling Terrestrial Environments for Bacterial Polyketides. Molecules, 22(5), 707. https://doi.org/10.3390/molecules22050707




