Chemical Composition of Laurencia obtusa Extract and Isolation of a New C15-Acetogenin
Abstract
:1. Introduction
2. Results
2.1. Chemical Composition of Laurencia obtusa Extract
2.1.1. Validation of 13C-NMR Method on Crude Extract
2.1.2. Application of 13C-NMR Method to Chromatography Fractions
2.2. Structure Elucidation of Compound 20
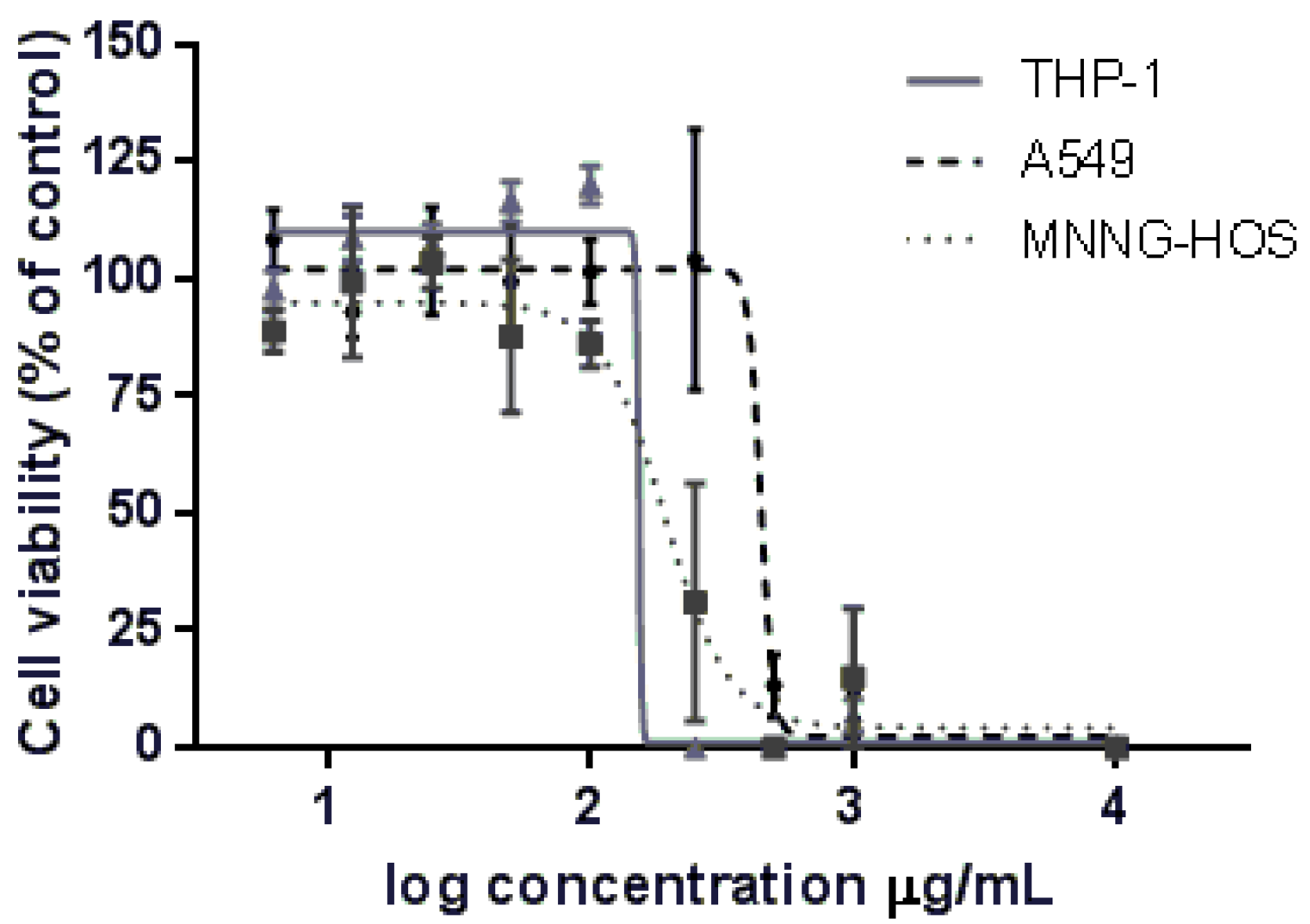
2.3. Cytotoxic Activity
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Extraction and Separation
4.3. NMR Analysis
4.4. Mass Spectrometry
4.5. Identification of the Components
4.6. Cell Culture
4.7. Assessment of Cell Viability
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, B.G.; Gloer, J.B.; Ji, N.Y.; Zhao, J.C. Halogenated organic molecules of rhodomelaceae origin: Chemistry and biology. Chem. Rev. 2013, 113, 3632–3685. [Google Scholar] [CrossRef] [PubMed]
- Harizani, M.; Ioannou, E.; Roussis, V. The Laurencia paradox: An endless source of chemiodiversity. Prog. Chem. Org. Nat. Prod. 2016, 102, 91–252. [Google Scholar]
- Lhullier, C.; Falkenberg, M.; Ioannou, E.; Quesada, A.; Papazafiri, P.; Horta, P.A.; Schenkel, E.P.; Vagias, C.; Roussis, V. Cytotoxic Halogenated Metabolites from the Brazilian Red Alga Laurencia catarinensis. J. Nat. Prod. 2010, 73, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Kladi, M.; Vagias, C.; Furnari, G.; Moreau, D.; Roussakis, C.; Roussis, V. Cytotoxic cuparene sesquiterpenes from Laurencia microcladia. Tetrahedron Lett. 2005, 46, 5723–5726. [Google Scholar] [CrossRef]
- Kladi, M.; Xenaki, H.; Vagias, C.; Papazafiri, P.; Roussis, V. New cytotoxic sesquiterpenes from the red algae Laurencia obtusa and Laurencia microcladia. Tetrahedron 2006, 62, 182–189. [Google Scholar] [CrossRef]
- Sutour, S.; Xu, T.; Casabianca, H.; Paoli, M.; de Rocca-Serra, D.; Tomi, F.; Garrido, M.; Pasqualini, V.; Aiello, A.; Castola, V.; et al. Chemical composition of extracts from Chaetomorpha linum (Miller) Kütz. A potential use in the cosmetic industry. Int. J. Phytocosmet. Nat. Ingred. 2015, 2, 5. [Google Scholar]
- Xu, T.; Sutour, S.; Casabianca, H.; Tomi, F.; Paoli, M.; Garrido, M.; Pasqualini, V.; Aiello, A.; Castola, V.; Bighelli, A. Rapid screenig of chemical compositions of Gracilia dura and Hypnea musciformis (Rhodophyta) from Corsican Lagoon. Int. J. Phytocosmet. Nat. Ingred. 2015, 2, 8. [Google Scholar]
- Tomi, F.; Casanova, J. 13C-NMR as a tool for identification of individual components of essential oils from Labiatae A review. Acta Hortic. 2006, 723, 185–192. [Google Scholar] [CrossRef]
- Bazzali, O.; Huy Thai, T.; Minh Hoi, T.; Sinh Khang, N.; Thi Hien, N.; Casanova, J.; Bighelli, A.; Tomi, F. Integrated analysis of the wood oil from Xanthocyparis vietnamensis Farjon & Hiep. by chromatographic and spectroscopic techniques. Molecules 2016, 21, 840. [Google Scholar]
- Rezzi, S.; Bighelli, A.; Castola, V.; Casanova, J. Direct identification and quantitative determination of acidic and neutral diterpenes using 13C-NMR spectroscopy. J. Appl. Spectrosc. 2002, 56, 312–317. [Google Scholar] [CrossRef]
- Blanc, M.C.; Bradesi, P.; Gonçalves, M.J.; Salgueiro, L.; Casanova, J. Essential oil of Dittrichia viscosa ssp. viscosa: Analysis by 13C-NMR and antimicrobial activity. Flavour Fragr. J. 2006, 21, 324–332. [Google Scholar] [CrossRef]
- Gonny, M.; Bradesi, P.; Casanova, J. Identification of the components of the essential oil from wild Corsican Daucus carota L. using 13C-NMR spectroscopy. Flavour Fragr. J. 2004, 19, 424–433. [Google Scholar] [CrossRef]
- Howard, B.M.; Fenical, W. α- and β-snyderol; new bromo-monocyclic sesquiterpenes from the seaweed Laurencia. Tetrahedron Lett. 1976, 1, 41–44. [Google Scholar] [CrossRef]
- Amico, V.; Caccamese, S.; Neri, P.; Russo, G.; Foti, M. Brasilane-type sesquiterpenoids from the Mediterranean red alga Laurencia obtusa. Phytochemistry 1991, 30, 1921–1927. [Google Scholar] [CrossRef]
- Stallard, M.O.; Fenical, W. The brasilenols, rearranged sesquiterpenes alcohols isolated from the marine opsthobranch Aplysia brasilana. Tetrahedron 1978, 34, 2077–2081. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Murphy, P.T.; Quinn, R.J.; Wells, R.J. New laurene derivatives from Laurencia filiformis. Aust. J. Chem. 1976, 29, 2533–2539. [Google Scholar] [CrossRef]
- Kadi, M.; Vagias, C.; Papazafiri, P.; Furnari, G.; Serio, D.; Roussis, V. New sesquiterpens from the red alga Laurencia microcladia. Tetrahedron 2007, 63, 7606–7611. [Google Scholar]
- Zhang, J.; Ding, L.P.; Liang, H.; Guo, X.Y.; Zhang, Q.Y. Sesquiterpenes from the red alga Laurencia tristicha. Biochem. Syst. Ecol. 2015, 60, 116–119. [Google Scholar] [CrossRef]
- König, G.M.; Wright, A.D. New C15-acetogenins and sesquiterpenes from the red alga Laurencia sp cf. L. gracilis. J. Nat. Prod. 1994, 57, 477–485. [Google Scholar]
- Suzuki, M.; Kurosawa, E. New aromatic sesquiterpenoids from the red alga Laurencia yamada (1). Tetrahedron Lett. 1978, 28, 2503–2506. [Google Scholar]
- Izac, R.R.; Sims, J.J. Marine Natural Products. 18. Iodinated sesquiterpenes from the red algal genus Laurencia. J. Am. Chem. Soc. 1979, 101, 6136–6137. [Google Scholar] [CrossRef]
- Boeckman, R.K., Jr.; Zhang, J.; Reeder, M.R. Synthetic and mechanistic studies of the retro-claisen rearrangement 4. an application to the total synthesis of (+)-laurenyne. Org. Lett. 2002, 4, 3891–3894. [Google Scholar] [CrossRef] [PubMed]
- Norte, M.; Gonzales, A.G.; Cataldo, F.; Rodriguez, M.L.; Brito, I. New examples of acyclic and cyclic C-15 actogenins from Laurencia pinnatifida. Reassignment of the absolute configuration for E and Z pinnatifidenyne. Tetrahedron 1991, 47, 9411–9418. [Google Scholar]
- Iliopoulou, D.; Vagias, C.; Harvala, C.; Roussis, V. C15-acetogenins from the red alga Laurencia obtusa. Phytochemistry 2002, 59, 111–116. [Google Scholar] [CrossRef]
- Norte, M.; Fernandez, J.J.; Cataldo, F.; Gonzales, A.G. E-dihydrorhodophytin, a C15-acetogenin from the red alga Laurencia pinnatifida. Phytochemistry 1989, 28, 647–649. [Google Scholar] [CrossRef]
- Guella, G.; Pietra, F. A new-skeleton diterpenoid, new prenylbisabolanes, and their putative biogenetic precursor, from the red seaweed Laurencia microcladia from II Rogiolo: Assigning the absolute configuration when two chiral halves are connected by single bonds. Helv. Chim. Acta 2000, 83, 2946–2952. [Google Scholar] [CrossRef]
- Howard, B.M.; Fenical, W. Obtusadiol, a unique bromoditerpenoid from the marine red alga Laurencia obtusa. Tetrahedron Lett. 1978, 28, 2453–2456. [Google Scholar] [CrossRef]
- Wright, J.L.C.; McInnes, A.G.; Shimizu, S.; Smith, D.G.; Walter, J.A.; Idler, D.; Khalil, W. Identification of C-24 alkyl epimers of marine sterols by 13C nuclear magnetic resonance spectroscopy. Can. J. Chem. 1978, 56, 1898–1903. [Google Scholar]
- McInnes, A.G.; Walter, J.A.; Wright, J.L.C. 13C-NMR spectra of Δ24(28) phytosterols. Magn. Reson. Chem. 1980, 13, 302–303. [Google Scholar] [CrossRef]
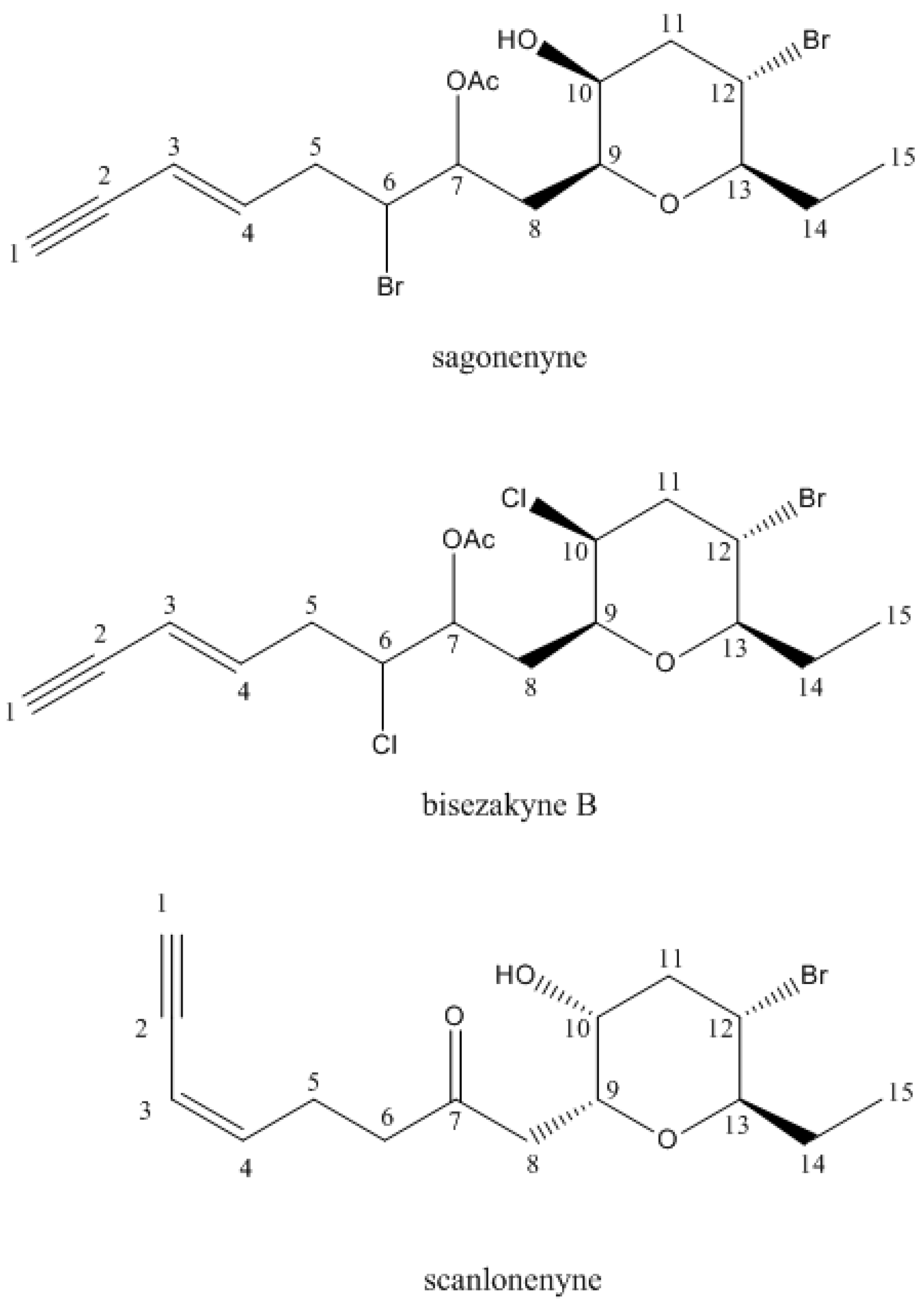
- Suzuki, M.; Nakano, S.; Takahashi, Y.; Abe, T.; Masuda, M. Bisezakyne-A and -B, halogenated C15-acetogenins from a Japanese Laurencia species. Phytochemistry 1991, 51, 657–662. [Google Scholar] [CrossRef]
- Suzuki, M.; Takahashi, Y.; Matsuo, Y.; Guiry, M.D.; Masuda, M. Scanlonenyne, a novel halogenated C15-acetogenin from the red alga Laurencia obtusa on Irish waters. Tetrahedron 1997, 12, 4271–4278. [Google Scholar] [CrossRef]
- Joulain, D.; König, W.A. The Atlas of Spectral Data of Sesquiterpenes Hydrocarbons; E.B.-Verlag: Hamburg, Germany, 1998. [Google Scholar]
- Wanke, T.; Phillippus, A.C.; Zatelli, G.A.; Vieira, L.F.O.; Lhullier, C.; Falkenberg, M. C15-acetogenins from the Laurencia complex: 50 years of research—An overview. Rev. Bras. Farmacogn. 2015, 25, 569–587. [Google Scholar] [CrossRef]
- Kornprobst, J.M. Substances Naturelles D’origine Marine; Lavoisier: Paris, France, 2005. [Google Scholar]
- Masuda, M.; Kogame, K.; Arisawa, S.; Suzuki, M. Morphology and halogenated secondary metabolites of three Gran Canaria species of Laurencia (Ceremiales, Rhodophyta). Bot. Mar. 1998, 41, 265–277. [Google Scholar] [CrossRef]
- Suzuki, M.; Vairappan, C.S. Halogenated secondary metabolites from Japanese species of the red algal gens Laurencia (Rhodomelaceae, Ceremiales). Curr. Top. Phytochem. 2005, 5, 1–38. [Google Scholar]
- Dellai, A.; Laajili, S.; Le Morvan, V.; Robert, J.; Bouraoui, A. Antiproliferative activity and phenolics of the Mediterranean seaweed Laurencia obtusa. Ind. Crops Prod. 2013, 47, 252–255. [Google Scholar] [CrossRef]
- Angawi, R.F.; Alarif, W.M.; Rehab, F.; BAdria, F.A.; Ayyad, S.E.N. New cytotoxic laurene-, cuparene-, and laurokamurene-type sesquiterpenes from the red alga Laurencia obtusa. Helv. Chim. Acta 2014, 97, 1388–1395. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |
| C | δ Extract (ppm) | δ Litt [13] (ppm) |
|---|---|---|
| 1 | 111.80 | 111.8 |
| 2 | 145.07 | 144.9 |
| 3 | 73.40 | 73.3 |
| 4 | 41.27 | 41.3 |
| 5 | 20.61 | 20.6 |
| 6 | 53.15 | 53.1 |
| 7 | 145.57 | 145.6 |
| 8 | 37.47 | 37.4 |
| 9 | 35.94 | 35.9 |
| 10 | 67.44 | 67.1 |
| 11 | 42.10 | 41.9 |
| 12 | 28.44 | 28.4 |
| 13 | 16.37 | 16.4 |
| 14 | 109.16 | 109.0 |
| 15 | 27.78 | 28.1 |
| N° | Components | NMR 3 | Overlap 4 | References |
|---|---|---|---|---|
| Sesquiterpenes | ||||
| 1 | β-snyderol 1 | 15/15 | 4 | [13] |
| 2 | α-snyderol 1 | 12/15 | 3 | [13] |
| 3 | Epibrasilenol 1 | 15/15 | 1 | [15] |
| 4 | Brasilenol 1 | 13/15 | 1 | [15] |
| 5 | 4-hydroxy-5-brasilene 1 | 15/15 | 1 | [14] |
| 6 | laurene | 15/15 | 0 | [16] |
| 7 | α-bromocuparene | 15/15 | 1 | [17] |
| 8 | α-isobromocuparene | 15/15 | 3 | [17] |
| 9 | laurinterol | 15/15 | 3 | [18] |
| 10 | iso-laurenisol | 15/15 | 0 | [19] |
| 11 | 3,7-dihydroxydihydrolaurene | 15/15 | 1 | [4] |
| 12 | laurane derivative | 15/15 | 0 | (compound 1 in [3]) |
| 13 | laurane derivative | 15/15 | 0 | (compound 10 in [20]) |
| 14 | 11-iodolaurinterol | 15/15 | 0 | [21] |
| C15-acetogenins | ||||
| 15 | 3-(E)-laurenyne | 14/15 | 0 | [22] |
| 16 | (Z) linear acetogenin | 15/15 | 2 | (compound 3 in [23]) |
| 17 | (E) linear acetogenin | 15/15 | 1 | (compound 4 in [23]) |
| 18 | 13-(E)-epipinnatifidenyne | 15/15 | 4 | [24] |
| 19 | (E)-dihydrorhodophytin | 15/15 | 5 | [25] |
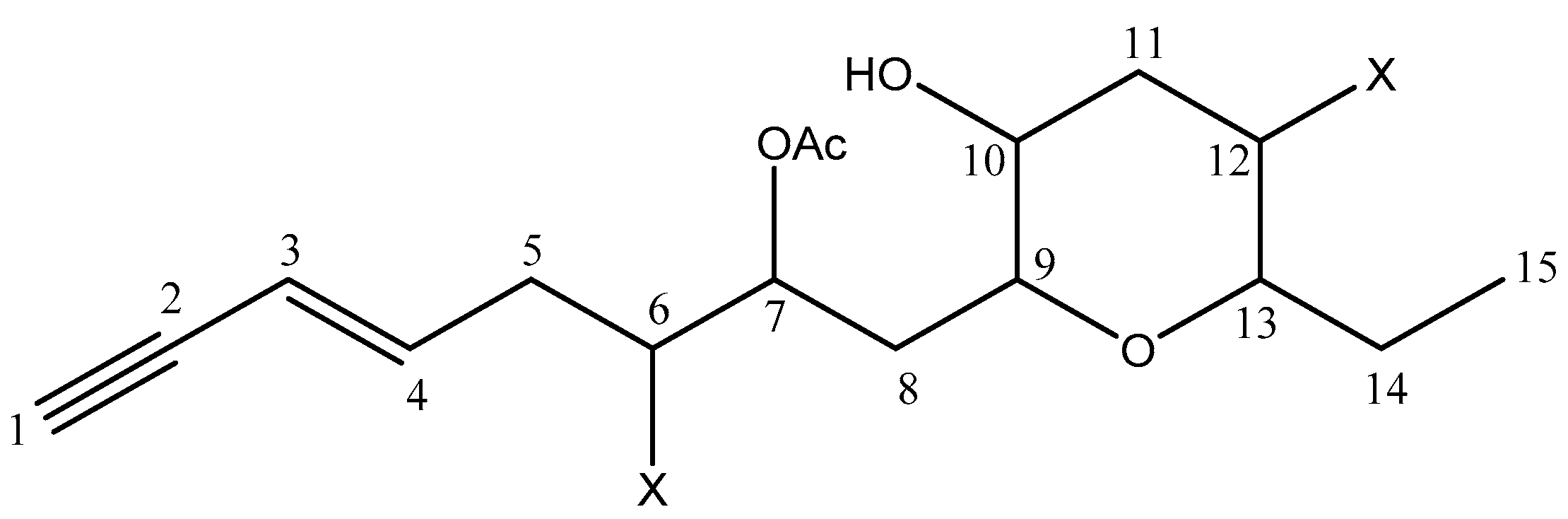
| 20 | Sagonenyne 2 | 15/15 | 0 | - |
| Diterpenes | ||||
| 21 | neorogioldiol | 20/20 | 3 | [26] |
| 22 | obtusadiol | 20/20 | 2 | [27] |
| Sterols | ||||
| 23 | fucosterol | 29/29 | 12 | [28] |
| 24 | cholesterol | 27/27 | 12 | [29] |
| C | δ 13C (ppm) | DEPT | δ 1H (ppm) | Multiplicity (J (Hz)) | COSY 1H–1H | HMBC H → C |
|---|---|---|---|---|---|---|
| 1 | 81.55 | CH | 2.85 | d (2.0) | C3; C4 | |
| 2 | 77.21 | C | - | - | ||
| 3 | 112.13 | CH | 5.55 | dd (15.9, 2.0) | H4; H1 | C2; C4; C5 |
| 4 | 141.29 | CH | 6.22 | dt (15.9, 7.1) | H4; H5a; H5b | C1; C3; C5; C6 |
| 5 | 38.77 | CH2 | a 2.55 | m | H6 | C3; C4; C6; C7 |
| b 2.70 | m | H6 | C2; C4; C6; C7 | |||
| 6 | 55.52 | CH | 4.04 | ddd (9.0, 4.9, 2.7) | H5a; H5b; H7 | C4; C5 |
| 7 | 71.38 | CH | 5.23 | dt (9.0, 2.7) | H8a; H8b; H6 | C5; C8; C9 |
| 8 | 35.35 | CH2 | a 1.78 | ddd (14.8, 9.0, 2.3) | H7 | C6 |
| b 2.04 | m | H9 | C9 | |||
| 9 | 76.25 | CH | 3.43 | ddd (10.7, 2.5, 1.0) | H8a; H8b; H10 | C8; C10; C13 |
| 10 | 69.95 | CH | 3.64 | br t | H11a; H11b | C12 |
| 11 | 43.42 | CH2 | a 2.12 | m | H10; H12 | C12; C13 |
| b 2.58 | m | H10; H12 | C9; C12; C13 | |||
| 12 | 47.50 | CH | 3.97 | ddd (12.3, 10.2, 4.8) | H11a; H11b; H13 | C11; C12; C14 |
| 13 | 83.98 | CH | 3.29 | ddd (10.4, 9.0, 2.5) | H14a; H14b; H12 | C9; C11; C12; C14; C15 |
| 14 | 26.35 | CH2 | a 1.48 | m | H13 | C12; C13; C15 |
| b 2.06 | m | H15 | C12; C13; C15 | |||
| 15 | 9.63 | CH3 | 0.99 | t (7.4) | H14a; H14b | C13; C14 |
| Ac | 20.87 | CH3 | 2.13 | s | ||
| 170.19 | C | - | - |
| Cell Lines | Laurencia obtusa |
|---|---|
| MNNG-HOS | 191.3 |
| THP-1 | 153.2 |
| A549 | 446.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esselin, H.; Sutour, S.; Liberal, J.; Cruz, M.T.; Salgueiro, L.; Siegler, B.; Freuze, I.; Castola, V.; Paoli, M.; Bighelli, A.; et al. Chemical Composition of Laurencia obtusa Extract and Isolation of a New C15-Acetogenin. Molecules 2017, 22, 779. https://doi.org/10.3390/molecules22050779
Esselin H, Sutour S, Liberal J, Cruz MT, Salgueiro L, Siegler B, Freuze I, Castola V, Paoli M, Bighelli A, et al. Chemical Composition of Laurencia obtusa Extract and Isolation of a New C15-Acetogenin. Molecules. 2017; 22(5):779. https://doi.org/10.3390/molecules22050779
Chicago/Turabian StyleEsselin, Hélène, Sylvain Sutour, Joana Liberal, Maria Teresa Cruz, Ligia Salgueiro, Benjamin Siegler, Ingrid Freuze, Vincent Castola, Mathieu Paoli, Ange Bighelli, and et al. 2017. "Chemical Composition of Laurencia obtusa Extract and Isolation of a New C15-Acetogenin" Molecules 22, no. 5: 779. https://doi.org/10.3390/molecules22050779







