Docosahexaenoic Acid Induces Expression of Heme Oxygenase-1 and NAD(P)H:quinone Oxidoreductase through Activation of Nrf2 in Human Mammary Epithelial Cells
Abstract
:1. Introduction
2. Results
2.1. DHA Upregulates Expression of HO-1 and NQO1
2.2. DHA Induces the Nuclear Translocation and Transcriptional Activity of Nrf2 through ARE Binding
2.3. DHA-Induced Expression of HO-1 and NQO1 Is Mediated by Nrf2
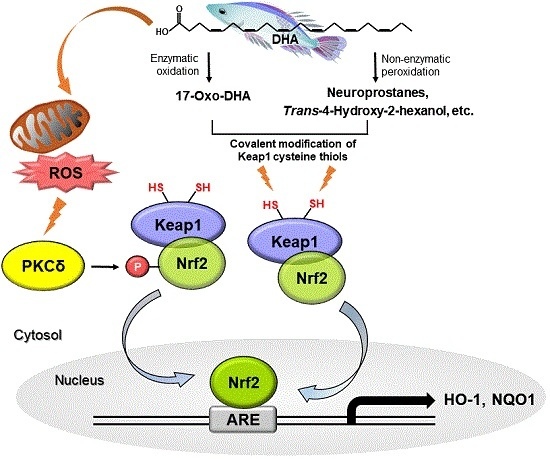
2.4. Cysteine Residues of Keap1 May Be Putative Targets of DHA for Its Induction of Nrf2-Driven Expression of HO-1 and NQO1
2.5. DHA-Derived Intracellular Reactive Oxygen Species (ROS) Accumulation Induces Phosphorylation of Nrf2 via the PKCδ Signaling
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Preparation and Culturing of MEF
4.4. Preparation of Nuclear Extracts
4.5. Western Blot Analysis
4.6. RT-PCR
4.7. ARE Luciferase Assay
4.8. Immunocytochemistry
4.9. ChIP Assay
4.10. Generation of Stable Cells Expressing Keap1 Constructs
4.11. BPM-Labeling Assay
4.12. Measurement of Intracellular Accumulation of ROS
4.13. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Otton, R.; Marin, D.P.; Bolin, A.P.; de Cassia Santos Macedo, R.; Campoio, T.R.; Fineto, C., Jr.; Guerra, B.A.; Leite, J.R.; Barros, M.P.; Mattei, R. Combined fish oil and astaxanthin supplementation modulates rat lymphocyte function. Eur. J. Nutr. 2012, 51, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.E.; Shahbakhti, H.; Azurdia, R.M.; Moison, R.M.; Steenwinkel, M.J.; Homburg, M.I.; Dean, M.P.; McArdle, F.; Beijersbergen van Henegouwen, G.M.; Epe, B.; et al. Effect of eicosapentaenoic acid, an omega-3 polyunsaturated fatty acid, on UVR-related cancer risk in humans. An assessment of early genotoxic markers. Carcinogenesis 2003, 24, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol. Nutr. Food Res. 2008, 52, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Bazan, N.G. Omega-3 fatty acids, pro-inflammatory signaling and neuroprotection. Curr. Opin. Clin. Nutr. 2007, 10, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Kwon, S.H.; Han, Y.M.; Hahm, K.B.; Kim, E.H. Omega-3 Polyunsaturated fatty acids as potential chemopreventive agents for gastric cancer. J. Cancer Prev. 2013, 18, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Wall, R.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C. Health implication of high dietary omega-6 polyunsaturated fatty acids. J. Nutr. Metab. 2012, 2012, 539426. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.D.; Whitley, B.M.; Hoyo, C.; Grant, D.J.; Iraggi, J.D.; Newman, K.A.; Gerber, L.; Taylor, L.A.; McKeever, M.G.; Freedland, S.J. A high ratio of dietary n-6/n-3 polyunsaturated fatty acids is associated with increased risk of prostate cancer. Nutr. Res. 2011, 31, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The omega-6/omega-3 fatty acid ratio, genetic variation, and cardiovascular disease. Asia Pac. J. Clin. Nutr. 2008, 17, 131–134. [Google Scholar] [PubMed]
- Black, H.S.; Thornby, J.I.; Gerguis, J.; Lenger, W. Influence of dietary omega-6, -3 fatty acid sources on the initiation and promotion stages of photocarcinogenesis. Photochem. Photobiol. 1992, 56, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.S.; Burill, C.; Rigotty, J. Effect of diets high in omega-3 and omega-6 fatty acids on initiation and postinitiation stages of colon carcinogenesis. Cancer Res. 1991, 51, 487–491. [Google Scholar] [PubMed]
- Huffman, S.L.; Harika, R.K.; Eilander, A.; Osendarp, S.J. Essential fats: How do they affect growth and development of infants and young children in developing countries? A literature review. Matern. Child Nutr. 2011, 7, 44–65. [Google Scholar] [CrossRef] [PubMed]
- Morland, S.L.; Martins, K.; Mazurak, V.C. n-3 polyunsaturated fatty acid supplementation during cancer chemotherapy. J. Nutr. Intermed. Metab. 2016, 5, 107–116. [Google Scholar] [CrossRef]
- Bougnoux, P.; Hajjaji, N.; Ferrasson, M.N.; Giraudeau, B.; Couet, C.; Le Floch, O. Improving outcome of chemotherapy of metastatic breast cancer by docosahexaenoic acid: A phase II trial. Br. J. Cancer 2009, 101, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.A.; Mourtzakis, M.; Chu, Q.; Baracos, V.E.; Reiman, T.; Mazurak, V.C. Supplementation with fish oil increases first-line chemotherapy efficacy in patients with advanced nonsmall cell lung cancer. Cancer 2011, 117, 3774–3780. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.D.; Tapper, A.R.; Zackert, W.E.; Yang, J.; Sanchez, S.C.; Montine, T.J.; Roberts, L.J. Formation of novel isoprostane-like compounds from docosahexaenoic acid. Adv. Exp. Med. Biol. 1999, 469, 343–347. [Google Scholar] [PubMed]
- De Smedt-Peyrusse, V.; Sargueil, F.; Moranis, A.; Harizi, H.; Mongrand, S.; Laye, S. Docosahexaenoic acid prevents lipopolysaccharide-induced cytokine production in microglial cells by inhibiting lipopolysaccharide receptor presentation but not its membrane subdomain localization. J. Neurochem. 2008, 105, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Tanaka, M.; Masuda, S.; Ohue-Kitano, R.; Yamakage, H.; Muranaka, K.; Wada, H.; Kusakabe, T.; Shimatsu, A.; Hasegawa, K.; et al. Omega-3 polyunsaturated fatty acids suppress the inflammatory responses of lipopolysaccharide-stimulated mouse microglia by activating SIRT1 pathways. Biochim. Biophys. Acta 2017, 1862, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Wiest, E.F.; Walsh-Wilcox, M.T.; Walker, M.K. Omega-3 polyunsaturated fatty acids protect against cigarette smoke-induced oxidative stress and vascular dysfunction. Toxicol. Sci. 2017, 156, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Lian, M.; Luo, W.; Sui, Y.; Li, Z.; Hua, J. Dietary n-3 PUFA protects mice from Con A induced liver injury by modulating regulatory T cells and PPAR-γ expression. PLoS ONE 2015, 10, e0132741. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.J.; Kundu, J.K.; Na, H.K.; Lee, J.S. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J. Nutr. 2005, 135, S2993–S3001. [Google Scholar]
- Surh, Y.J.; Kundu, J.K.; Na, H.K. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008, 74, 1526–1539. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Johnson, J.A. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J. Biochem. Mol. Biol. 2004, 37, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.J.; Na, H.K.; Park, J.M.; Lee, H.N.; Kim, W.; Yoon, I.S.; Kim, D.D. 15-Deoxy-Δ12,14-prostaglandin J2, an electrophilic lipid mediator of anti-inflammatory and pro-resolving signaling. Biochem. Pharmacol. 2011, 82, 1335–1351. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Saito, Y.; Yoshida, Y.; Sekine, A.; Noguchi, N.; Niki, E. 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J. Biol. Chem. 2005, 280, 41921–41927. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, J.; Sekhar, K.R.; Yin, H.; Yared, N.F.; Schneider, S.N.; Sasi, S.; Dalton, T.P.; Anderson, M.E.; Chan, J.Y.; et al. Novel n-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between Keap1 and Cullin3. J. Biol. Chem. 2007, 282, 2529–2537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kang, M.I.; Watai, Y.; Tong, K.I.; Shibata, T.; Uchida, K.; Yamamoto, M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 2006, 26, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed]
- Niture, S.K.; Jain, A.K.; Jaiswal, A.K. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J. Cell Sci. 2009, 122, 4452–4464. [Google Scholar] [CrossRef] [PubMed]
- Numazawa, S.; Ishikawa, M.; Yoshida, A.; Tanaka, S.; Yoshida, T. Atypical protein kinase C mediates activation of NF-E2-related factor 2 in response to oxidative stress. Am. J. Physiol. Cell Physiol. 2003, 285, C334–C342. [Google Scholar] [CrossRef] [PubMed]
- Kundu, J.K.; Surh, Y.J. Nrf2-Keap1 signaling as a potential target for chemoprevention of inflammation-associated carcinogenesis. Pharm. Res. 2010, 27, 999–1013. [Google Scholar] [CrossRef] [PubMed]
- Tkachev, V.O.; Menshchikova, E.B.; Zenkov, N.K. Mechanism of the Nrf2/Keap1/ARE signaling system. Biochemistry (Moscow) 2011, 76, 407–422. [Google Scholar] [CrossRef]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J.; Bazan, N.G. Docosahexaenoic acid and the aging brain. J. Nutr. 2008, 138, 2510–2514. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 12475–12480. [Google Scholar] [CrossRef] [PubMed]
- Rose, D.P.; Connolly, J.M. Omega-3 fatty acids as cancer chemopreventive agents. Pharmacol. Ther. 1999, 83, 217–244. [Google Scholar] [CrossRef]
- Chen, C.Y.; Jang, J.H.; Li, M.H.; Surh, Y.J. Resveratrol upregulates heme oxygenase-1 expression via activation of NF-E2-related factor 2 in PC12 cells. Biochem. Biophys. Res. Commun. 2005, 331, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Hu, B.; Cederbaum, A.I. Diallyl sulfide induces heme oxygenase-1 through MAPK pathway. Arch. Biochem. Biophys. 2004, 432, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Q.; Kim, M.Y.; Godoy, L.C.; Thiantanawat, A.; Trudel, L.J.; Wogan, G.N. Nitric oxide activation of Keap1/Nrf2 signaling in human colon carcinoma cells. Proc. Natl. Acad. Sci. USA 2009, 106, 14547–14551. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Jang, J.H.; Chen, C.Y.; Na, H.K.; Surh, Y.J. A formulated red ginseng extract rescues PC12 cells from PCB-induced oxidative cell death through Nrf2-mediated upregulation of heme oxygenase-1 and glutamate cysteine ligase. Toxicology 2010, 278, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Surh, Y.J. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005, 224, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Wakabayashi, N.; Kensler, T.W. Chemoprevention through the Keap1-Nrf2 signaling pathway by phase 2 enzyme inducers. Mutat. Res. 2004, 555, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.M.; Yin, M.C. EPA or DHA enhanced oxidative stress and aging protein expression in brain of d-galactose treated mice. Biomedicine (Taipei) 2016, 6, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, M.S.; Scolamacchia, M.; Betancor, M.; Roo, J.; Caballero, M.J.; Terova, G.; Witten, P.E. Effects of dietary DHA and α-tocopherol on bone development, early mineralisation and oxidative stress in Sparus aurata (Linnaeus, 1758) larvae. Br. J. Nutr. 2013, 109, 1796–1805. [Google Scholar] [CrossRef] [PubMed]
- Roche, E.; Ramírez-Tortosa, C.L.; Arribas, M.I.; Ochoa, J.J.; Sirvent-Belando, J.E.; Battino, M.; Ramírez-Tortosa, M.C.; González-Alonso, A.; Pérez-López, M.P.; Quiles, J.L. Comparative analysis of pancreatic changes in aged rats fed life long with sunflower, fish, or olive oils. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yamamoto, M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Giudice, A.; Arra, C.; Turco, M.C. Review of molecular mechanisms involved in the activation of the Nrf2-ARE signaling pathway by chemopreventive agents. Methods Mol. Biol. 2010, 647, 37–74. [Google Scholar] [PubMed]
- Lee, J.H.; Khor, T.O.; Shu, L.; Su, Z.Y.; Fuentes, F.; Kong, A.N. Dietary phytochemicals and cancer prevention: Nrf2 signaling, epigenetics, and cell death mechanisms in blocking cancer initiation and progression. Pharmacol. Ther. 2013, 137, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Fujimoto, K.; Miyazawa, T. Polyunsaturated (n-3) fatty acids susceptible to peroxidation are increased in plasma and tissue lipids of rats fed docosahexaenoic acid-containing oils. J. Nutr. 2000, 130, 3028–3033. [Google Scholar] [PubMed]
- Majkova, Z.; Layne, J.; Sunkara, M.; Morris, A.J.; Toborek, M.; Hennig, B. Omega-3 fatty acid oxidation products prevent vascular endothelial cell activation by coplanar polychlorinated biphenyls. Toxicol. Appl. Pharmacol. 2011, 251, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Ishikado, A.; Morino, K.; Nishio, Y.; Nakagawa, F.; Mukose, A.; Sono, Y.; Yoshioka, N.; Kondo, K.; Sekine, O.; Yoshizaki, T.; et al. 4-Hydroxy hexenal derived from docosahexaenoic acid protects endothelial cells via Nrf2 activation. PLoS ONE 2013, 8, e69415. [Google Scholar] [CrossRef] [PubMed]
- Nagayama, K.; Morino, K.; Sekine, O.; Nakagawa, F.; Ishikado, A.; Iwasaki, H.; Okada, T.; Tawa, M.; Sato, D.; Imamura, T.; et al. Duality of n-3 polyunsaturated fatty acids on Mcp-1 expression in vascular smooth muscle: A potential role of 4-hydroxy hexenal. Nutrients 2015, 7, 8112–8126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, S.; Mao, L.; Leak, R.K.; Shi, Y.; Zhang, W.; Hu, X.; Sun, B.; Cao, G.; Gao, Y.; et al. Omega-3 fatty acids protect the brain against ischemic injury by activating Nrf2 and upregulating heme oxygenase 1. J. Neurosci. 2014, 34, 1903–1915. [Google Scholar] [CrossRef] [PubMed]
- Papaiahgari, S.; Kleeberger, S.R.; Cho, H.Y.; Kalvakolanu, D.V.; Reddy, S.P. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J. Biol. Chem. 2004, 279, 42302–42312. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Huang, Z.; Zhang, D.D. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS ONE 2009, 4, e6588. [Google Scholar] [CrossRef] [PubMed]
- Gjyshi, O.; Flaherty, S.; Veettil, M.V.; Johnson, K.E.; Chandran, B.; Bottero, V. Kaposi’s sarcoma-associated herpesvirus induces Nrf2 activation in latently infected endothelial cells through SQSTM1 phosphorylation and interaction with polyubiquitinated Keap1. J. Virol. 2015, 89, 2268–2286. [Google Scholar] [CrossRef] [PubMed]
- Terrill, J.R.; Radley-Crabb, H.G.; Grounds, M.D.; Arthur, P.G. N-Acetylcysteine treatment of dystrophic mdx mice results in protein thiol modifications and inhibition of exercise induced myofibre necrosis. Neuromuscul. Disord. 2011, 22, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, H.G.; Park, S.A.; Kundu, J.K.; Keum, Y.S.; Cha, Y.N.; Na, H.K.; Surh, Y.J. Keap1 cysteine 288 as a potential target for diallyl trisulfide-induced Nrf2 activation. PLoS ONE 2014, 9, e85984. [Google Scholar] [CrossRef] [PubMed]
- Park, S.A.; Lee, M.H.; Na, H.K.; Surh, Y.J. 4-Hydroxyestradiol induces mammary epithelial cell transformation through Nrf2-mediated heme oxygenase-1 overexpression. Oncotarget 2017, 8, 164–178. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bang, H.-Y.; Park, S.-A.; Saeidi, S.; Na, H.-K.; Surh, Y.-J. Docosahexaenoic Acid Induces Expression of Heme Oxygenase-1 and NAD(P)H:quinone Oxidoreductase through Activation of Nrf2 in Human Mammary Epithelial Cells. Molecules 2017, 22, 969. https://doi.org/10.3390/molecules22060969
Bang H-Y, Park S-A, Saeidi S, Na H-K, Surh Y-J. Docosahexaenoic Acid Induces Expression of Heme Oxygenase-1 and NAD(P)H:quinone Oxidoreductase through Activation of Nrf2 in Human Mammary Epithelial Cells. Molecules. 2017; 22(6):969. https://doi.org/10.3390/molecules22060969
Chicago/Turabian StyleBang, Hye-Yoon, Sin-Aye Park, Soma Saeidi, Hye-Kyung Na, and Young-Joon Surh. 2017. "Docosahexaenoic Acid Induces Expression of Heme Oxygenase-1 and NAD(P)H:quinone Oxidoreductase through Activation of Nrf2 in Human Mammary Epithelial Cells" Molecules 22, no. 6: 969. https://doi.org/10.3390/molecules22060969