Improved Method for Reliable HMW-GS Identification by RP-HPLC and SDS-PAGE in Common Wheat Cultivars
Abstract
:1. Introduction
2. Results and Discussion
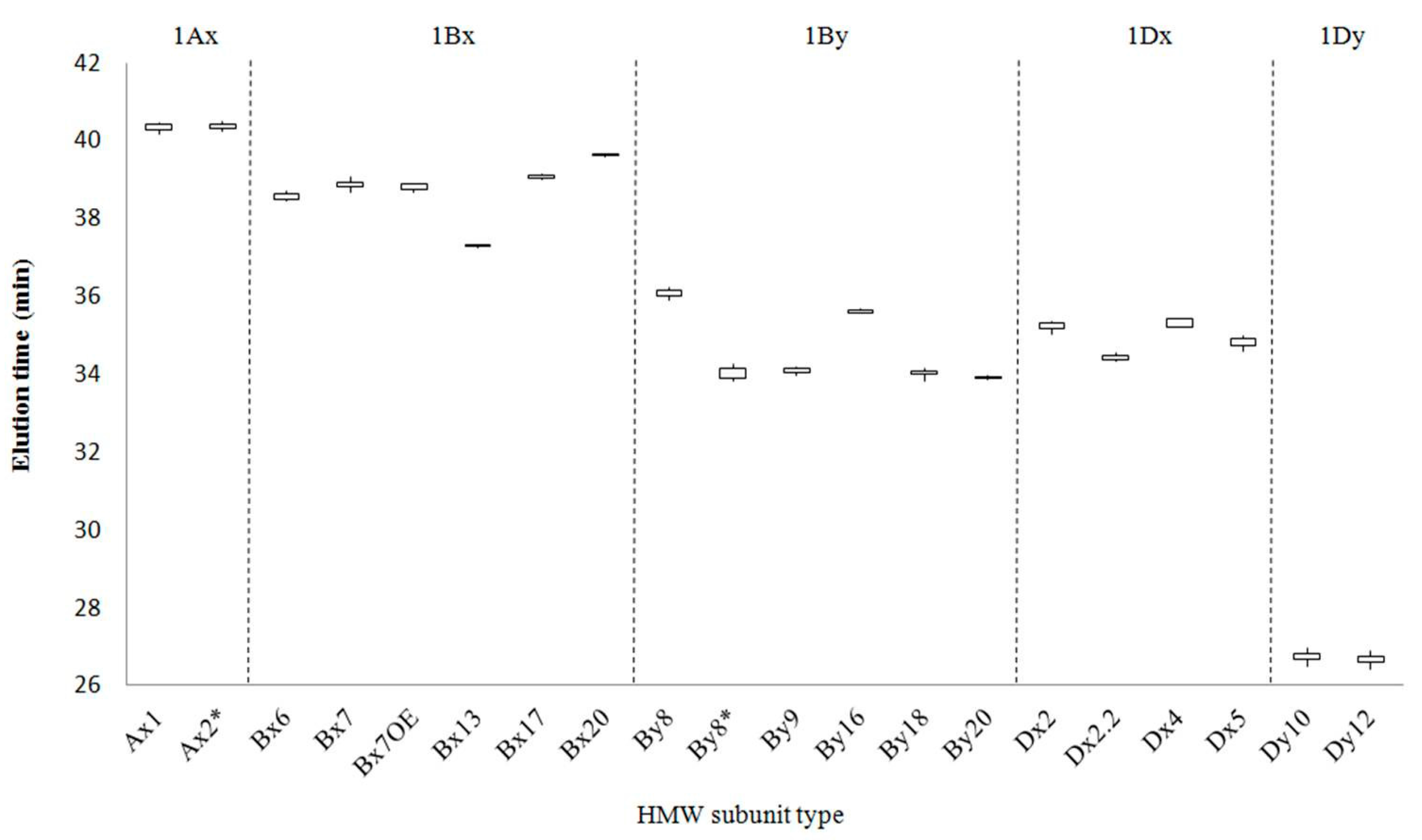
2.1. Identification of HMW-GS by both RP-HPLC and SDS-PAGE in Standard Cultivars
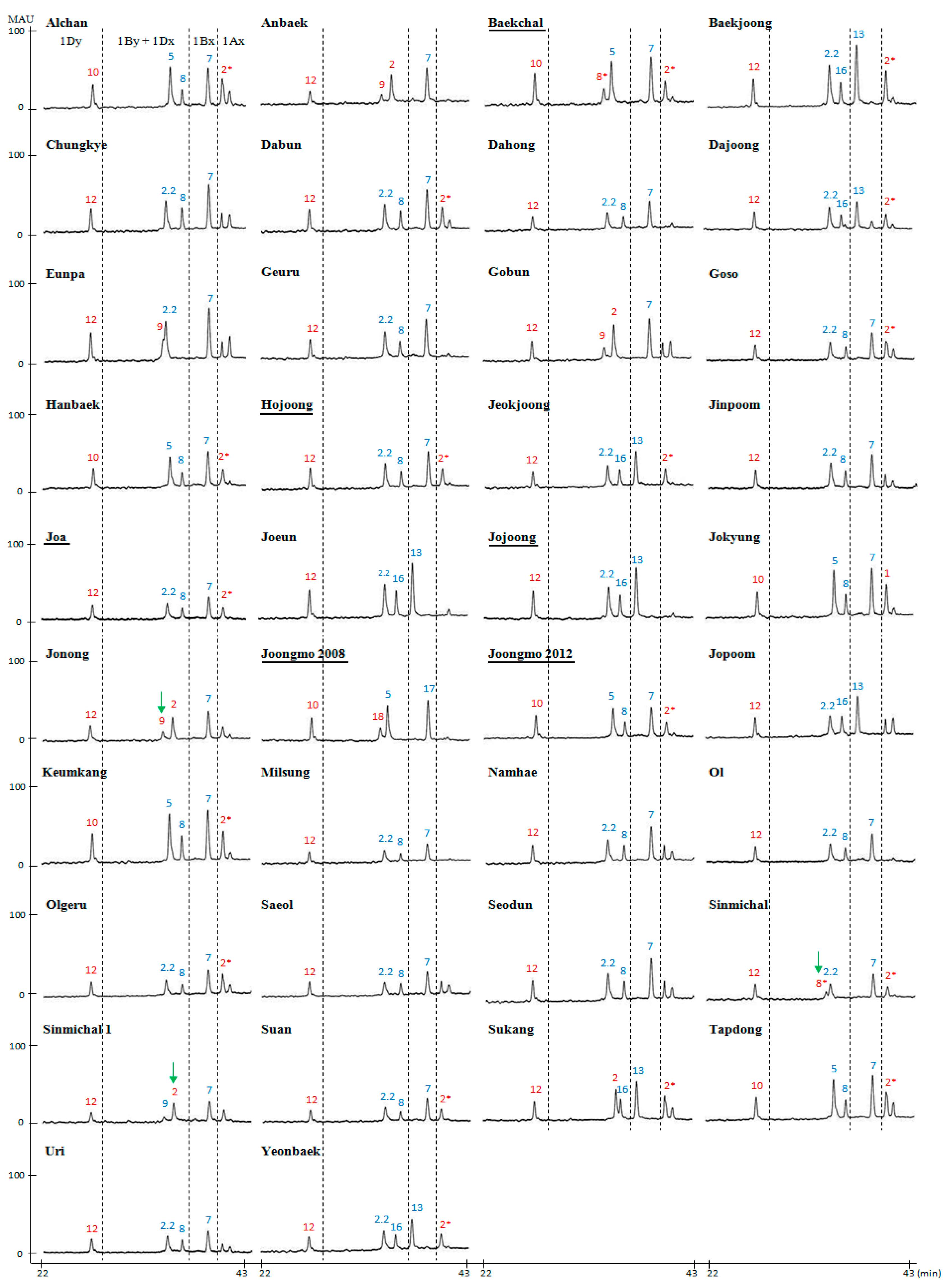
2.2. Compositional Analysis of HMW-GS in 38 Korean Wheat Cultivars
3. Experimental Section
3.1. Plant Materials
3.2. Glutenin Extraction
3.3. Analysis of the Individual HMW-GS Using RP-HPLC
3.4. SDS-PAGE
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Branlard, G.; Dardevet, M. Diversity of grain protein and bread wheat quality. II. Correlation between high molecular weight subunits of glutenin and flour quality characteristics. J. Cereal Sci. 1985, 3, 345–354. [Google Scholar] [CrossRef]
- Barro, F.; Rooke, L.; Bekes, F.; Gras, P.; Tatham, A.S.; Fido, R.; Lazzeri, P.A.; Shewry, P.R.; Barcelo, P. Transformation of wheat with HMW subunit genes results in improved functional properties. Nat. Biotechnol. 1997, 15, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, C.W. Biopolymers-Giant proteins with flour power. Nature 1996, 381, 738–739. [Google Scholar] [CrossRef] [PubMed]
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on breadmaking quality. Annu. Rev. Plant Physiol. 1987, 38, 141–153. [Google Scholar] [CrossRef]
- Payne, P.I.; Law, C.N.; Mudd, E.E. Control by homologous group 1 chromosomes of the high-molecular-weight subunits of glutenin, a major protein of wheat endosperm. Theor. Appl. Genet. 1980, 58, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G.; Tatham, A.S. High molecular weight subunits of wheat glutenin. J. Cereal Sci. 1992, 15, 105–120. [Google Scholar] [CrossRef]
- Payne, P.I.; Lawrence, G.J. Catalogue of alleles for the complex gene loci Glu-A1, Glu-B1 and Glu-D1 which code for high-molecular weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 1983, 11, 29–35. [Google Scholar]
- Gianibelli, M.C.; Larroque, O.R.; MacRitchie, F.; Wrigley, C.W. Biochemical, genetic, and molecular characterization of wheat glutenin and its component subunits. Cereal Chem. 2001, 78, 635–646. [Google Scholar] [CrossRef]
- Vawser, M.J.; Cornish, G.B. Over-expression of HMW glutenin subunit Glu-B1 7x in hexaploid wheat varieties (Triticum aestivum). Crop Pasture Sci. 2004, 55, 577–588. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Gao, X.; Wang, K.; Wang, S.; Zhang, Y.; He, Z.; Ma, W.; Yan, Y. Comparative proteome analysis of glutenin synthesis and accumulation in developing grains between superior and poor quality bread wheat cultivars. J. Sci. Food Agric. 2012, 92, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Butow, B.J.; Ma, W.; Gale, K.R.; Cornish, G.B.; Rampling, L.; Larroque, O.; Morell, M.K.; Bekes, F. Molecular discrimination of Bx7 alleles demonstrates that a highly expressed high molecular weight glutenin allele has a major impact on wheat flour dough strength. Theor. Appl. Genet. 2003, 107, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- D’Ovidio, R.; Masci, S.; Porceddu, E.; Kasarda, D. Duplication of the high molecular weight glutenin subunit gene in bread wheat (Triticum aestivum L.) cultivar ‘Red River 68’. Plant Breed. 1997, 116, 525–531. [Google Scholar] [CrossRef]
- Radovanovic, N.; Cloutier, S.; Brown, D.; Humphreys, D.G.; Lukow, O.M. Genetic variance for gluten strength contributed by high molecular weight glutenin proteins. Cereal Chem. 2002, 79, 843–849. [Google Scholar] [CrossRef]
- An, X.; Zhang, Q.; Yan, Y.; Li, Q.; Zhang, Y.; Wang, A.; Pei, Y.; Tian, J.; Wang, H.; Hsam, S.L.K.; et al. Cloning and molecular characterization of three novel LMW-i glutenin subunit genes from cultivated einkorn (Triticum monococcum L.). Theor. Appl. Genet 2006, 113, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Bietz, J.A. Separation of cereal proteins by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1983, 255, 219–238. [Google Scholar] [CrossRef]
- Courcoux, P.; Serot, T.; Larre, C.; Popineau, Y. Characterization and identification of wheat cultivars by multi-dimensional analysis of reversed-phase high-performance liquid chromatograms. J. Chromatogr. A 1992, 596, 225–232. [Google Scholar] [CrossRef]
- Cozzolino, R.; Giorgi, S.D.; Fisichella, S.; Garozzo, D.; Lafiandra, D.; Palermo, A. Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of high molecular weight glutenin subunits 1Bx7 and 1Dy10 in Cheyenne cultivar. Rapid Commun. Mass Spectrom. 2001, 15, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Hao, C.; Wang, A.; Cai, M.; Yan, Y. Characterization of HMW glutenin subunits in bread and tetraploid wheats by reversed-phase high-performance liquid chromatography. Cereal Res. Commun. 2009, 37, 65–73. [Google Scholar] [CrossRef]
- Gao, L.; Ma, W.; Chen, J.; Wang, K.; Li, J.; Wang, S.; Bekes, F.; Appels, R.; Yan, Y. Characterization and comparative analysis of wheat high molecular weight glutenin subunits by SDS-PAGE, RP-HPLC, HPCE, and MALDI-TOF-MS. J. Agric. Food Chem. 2010, 58, 2777–2786. [Google Scholar] [CrossRef] [PubMed]
- Naeem, H.A.; Sapirstein, H.D. Ultra-fast separation of wheat glutenin subunits by reversed-phase HPLC using a superficially porous silica-based column. J. Cereal Sci. 2007, 46, 157–168. [Google Scholar] [CrossRef]
- Yan, X.; Liu, W.; Yu, Z.; Han, C.; Zeller, F.J.; Hsam, S.L.K.; Yan, Y. Rapid separation and identification of wheat HMW glutenin subunits by UPLC and comparative analysis with HPLC. Aust. J. Crop Sci. 2014, 140–147. [Google Scholar]
- Visioli, G.; Comastri, A.; Imperiale, D.; Paredi, G.; Faccini, A.; Marmiroli, N. Gel-based and gel-free analytical methods for the detection of HMW-GS and LMW-GS in wheat flour. Food Anal. Methods 2016, 9, 469–476. [Google Scholar] [CrossRef]
- Liu, L.; Wang, A.; Appels, R.; Ma, J.; Xia, X.; Lan, P.; He, Z.; Bekes, F.; Yan, Y.; Ma, W. A MALDI-TOF based analysis of high molecular weight glutenin subunits for wheat breeding. J. Cereal Sci. 2009, 50, 295–301. [Google Scholar] [CrossRef]
- Dupont, F.M.; Chan, R.; Lopez, R. Molar fractions of high-molecular-weight glutenin subunits are stable when wheat is grown under various mineral nutrition and temperature regimens. J. Cereal Sci. 2007, 45, 134–139. [Google Scholar] [CrossRef]
- Branlard, G.; Dardevet, M.; Amiour, N.; Igrejas, G. Allelic diversity of HMW and LMW glutenin subunits and omega-gliadins in French bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 2003, 50, 669–679. [Google Scholar] [CrossRef]
- Hua, C.; Takata, K.; Yang-Fen, Z.; Ikeda, T.M.; Yanaka, M.; Nagamine, T.; Fujimaki, H. Novel high molecular weight glutenin subunits at the Glu-D1 locus in wheat landraces from the Xinjiang District of China and relationship with winter habit. Breed. Sci. 2005, 55, 459–463. [Google Scholar] [CrossRef]
- Park, C.S.; Kang, C.S.; Jeung, J.U.; Woo, S.H. Influence of allelic variations in glutenin on the quality of pan bread and white salted noodles made from Korean wheat cultivars. Euphytica 2011, 180, 235–250. [Google Scholar] [CrossRef]
- Shin, S.; Kang, C.S.; Kim, K.H.; Park, C.S. Analysis of Glutenin Compositions in Korean Wheat Cultivar Using SDS-PAGE and PCR. Korean J. Breed. Sci. 2012, 44. [Google Scholar]
- Singh, N.K.; Sheperd, K.W.; Cornish, G.B. A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J. Cereal Sci. 1991, 14, 203–208. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, Y.T.; Kim, H.J.; Lee, J.H.; Kang, C.S.; Lim, S.H.; Ha, S.H.; Ahn, S.N.; Kim, Y.M. Characterization of the HMW-GS 1Dx2.2 gene and its protein in a common Korean wheat variety. SABRAO J. Breed. Genet. 2013, 45, 159–168. [Google Scholar]
Sample Availability: Samples are not available from the authors. |
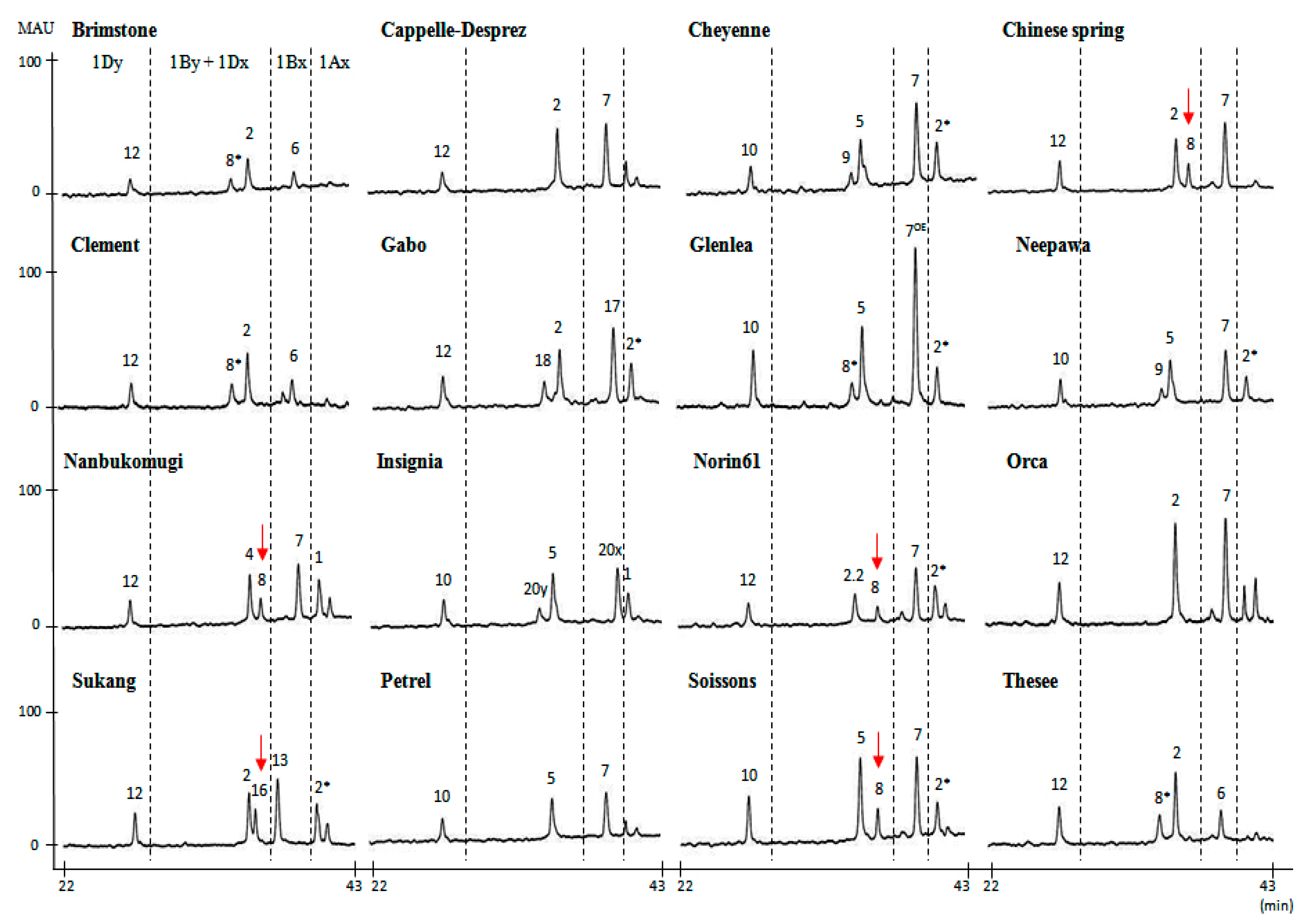
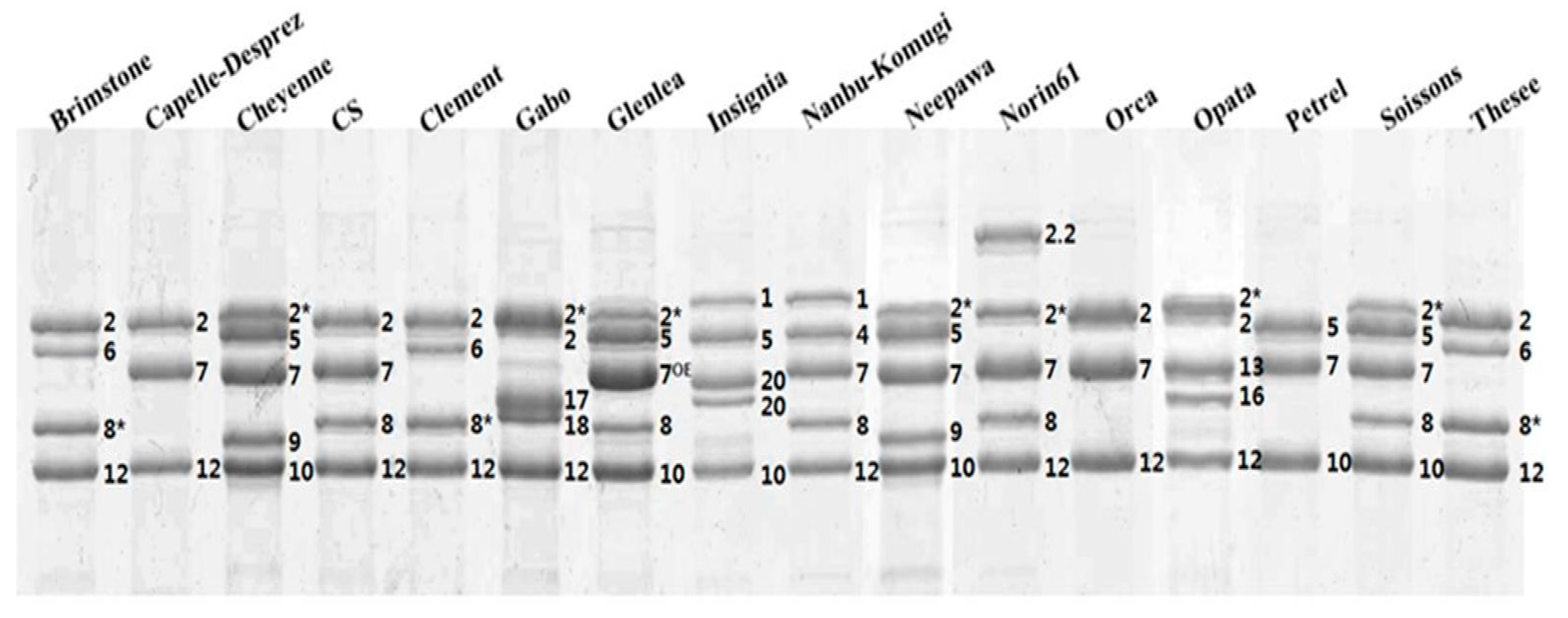

| Cultivar | HMW-GS | Reference | ||
|---|---|---|---|---|
| Glu-A1 | Glu-B1 | Glu-D1 | ||
| Brimstone | N | 6 + 8* | 2 + 12 | Liu et al. [24] |
| Cappelle-Desprez | N | 7 | 2 + 12 | Liu et al. [24] |
| Cheyenne | 2* | 7 + 9 | 5 + 10 | Dupont et al. [25] |
| Chinese Spring | N | 7 + 8 | 2 + 12 | Liu et al. [24] |
| Clement | N | 6 + 8* | 2 + 12 | Liu et al. [24] |
| Gabo | 2* | 17 + 18 | 2 + 12 | Liu et al. [24] |
| Glenlea | 2* | 7OE + 8* | 5 + 10 | Naeem and Sapirstein [21] |
| Insignia | 1 | 20 + 20 | 5 + 10 | Branlard [26] |
| Nanbu-komugi | 1 | 7 + 8 | 4 + 12 | Liu et al. [24] |
| Neepwa | 2* | 7 + 9 | 5 + 10 | Liu et al. [24] |
| Norin61 | 2 * | 7 + 8 | 2.2 + 12 | Hua et al. [27] |
| Orca | N | 7 | 2 + 12 | Liu et al. [24] |
| Sukang a | 2* | 13 + 16 | 2 + 12 | Park et al. [28] |
| Petrel | N | 7 | 5 + 10 | Liu et al. [24] |
| Soissons | 2* | 7 + 8 | 5 + 10 | Liu et al. [24] |
| Thesee | N | 6 + 8* | 2 + 12 | Liu et al. [24] |
| Type | HMW-GS | Number of Cultivars | Total Number of Analyses | Average Retention Time (min) | RSD (%) |
|---|---|---|---|---|---|
| Ax | Null | 7 | |||
| 1 | 2 | 9 | 40.307 ± 0.078 | 0.195 | |
| 2* | 7 | 18 | 40.354 ± 0.077 | 0.190 | |
| Bx | 6 | 3 | 8 | 38.516 ± 0.059 | 0.154 |
| 7 | 9 | 25 | 38.842 ± 0.077 | 0.199 | |
| 7OE | 1 | 6 | 38.770 ± 0.078 | 0.201 | |
| 13 | 1 | 3 | 37.285 ± 0.030 | 0.081 | |
| 17 | 1 | 5 | 39.075 ± 0.053 | 0.138 | |
| 20x | 1 | 4 | 39.603 ± 0.061 | 0.155 | |
| By | 8 | 4 | 12 | 36.060 ± 0.083 | 0.231 |
| 8* | 4 | 10 | 33.948 ± 0.098 | 0.290 | |
| 9 | 2 | 5 | 34.091 ± 0.085 | 0.252 | |
| 16 | 1 | 3 | 35.604 ± 0.039 | 0.109 | |
| 18 | 1 | 5 | 34.022 ± 0.108 | 0.319 | |
| 20y | 1 | 3 | 33.894 ± 0.043 | 0.127 | |
| Dx | 2 | 8 | 24 | 35.195 ± 0.114 | 0.325 |
| 2.2 | 1 | 3 | 34.428 ± 0.082 | 0.238 | |
| 4 | 1 | 3 | 35.269 ± 0.094 | 0.267 | |
| 5 | 6 | 17 | 34.792 ± 0.119 | 0.343 | |
| Dy | 10 | 6 | 10 | 26.736 ± 0.101 | 0.377 |
| 12 | 10 | 18 | 26.528 ± 0.098 | 0.370 |
| Cultivar | Pedigree | HMW-GS | ||
|---|---|---|---|---|
| Glu-A1 | Glu-B1 | Glu-D1 | ||
| Alchan | Suwon210/Tapdong | 2* | 7 + 8 | 5 + 10 |
| Anbaek | Sae/Geuru | N | 7 + 9 | 2 + 12 |
| Baekchal a | Sinmichal /Keumkang | 2* | 7 + 8* | 5 + 10 |
| Baekjoong | Keumkang/Olgeuru | 2* | 13 + 16 | 2.2 + 12 |
| Chungkye | Norin4/Sharbatisonora | N | 7 + 8 | 2.2 + 12 |
| Dabun | Suwon234//sw76039/Suwon220/3/Keumkang | 2* | 7 + 8 | 2.2 + 12 |
| Dahong | Norin72/Wonkwang | N | 7 + 8 | 2.2 + 12 |
| Dajoong | SW992114-NM-131-7/Gobun | 2* | 13 + 16 | 2.2 + 12 |
| Eunpa | Chugoku81/3/Tob-CNO//Yuksung3/Suwon185 | N | 7 + 9 | 2.2 + 12 |
| Geuru | Strampelli/69D-3607//Chokwang | N | 7 + 8 | 2.2 + 12 |
| Gobun | Eunpa/Tapdong//Eunpa/Shannung6521 | N | 7 + 9 | 2 + 12 |
| Goso | Gobun/Ol | 2* | 7 + 8 | 2.2 + 12 |
| Hanbaek | Shann7859/Keumkang//Guamuehill | 2* | 7 + 8 | 5 + 10 |
| Hojoong a | Alchan *2/3/Chunm18//JUP/BJY/4/Keumkang | 2* | 7 + 8 | 2.2 + 12 |
| Jeokjoong | Keumkang/Tapdong | 2* | 13 + 16 | 2.2 + 12 |
| Jinpoom | Geuru/Genaro81 | N | 7 + 8 | 2.2 + 12 |
| Joa a | SW86054-MB-27-3-2-1-1-1/Sumai#3 | 2* | 7 + 8 | 2.2 + 12 |
| Joeun | Eunpa/Suwon242 | N | 13 + 16 | 2.2 + 12 |
| Jojoong a | Suwon272/Olgeuru//Keumkang/Suwon252 | N | 13 + 16 | 2.2 + 12 |
| Jokyung | Seri82/Keumkang | 1 | 7 + 8 | 5 + 10 |
| Jonong | Suwon234/SW80199 | N | 7 + 9 | 2 + 12 |
| Joongmo2008 a | Eunpa *2//SH3/CBRD/3/Keumkang | N | 17 + 18 | 5 + 10 |
| Joongmo2012 a | Sinmichal/Keumkang | 2* | 7 + 8 | 5 + 10 |
| Jopoom | Kanto75//OR8500494P/Bezostaya | N | 13 + 16 | 2.2 + 12 |
| Keumkang | Geuru/Kanto75//Eunpa | 2* | 7 + 8 | 5 + 10 |
| Milsung | Shirogane//Norin43/Sonalika | N | 7 + 8 | 2.2 + 12 |
| Namhae | Ol/Calidad | N | 7 + 8 | 2.2 + 12 |
| Ol | Norin72/Norin12 | N | 7 + 8 | 2.2 + 12 |
| Olgeuru | Geuru/Chokwang//Saikai143 | 2* | 7 + 8 | 2.2 + 12 |
| Saeol | Shirogane//Norin43/Sonalika | N | 7 + 8 | 2.2 + 12 |
| Seodun | Geuru/Genaro81 | N | 7 + 8 | 2.2 + 12 |
| Sinmichal | Olgeuru//Kanto107/BaiHuo | 2* | 7 + 8* | 2.2 + 12 |
| Sinmichal1 | Alchan//Kanto107/BaiHuo | N | 7 + 9 | 2 + 12 |
| Suan | Keumkang/Eunpa//Keumkang | 2* | 7 + 8 | 2.2 + 12 |
| Sukang | Suwon266/Asakaze | 2* | 13 + 16 | 2 + 12 |
| Tapdong | Chugoku81//Suwon158/Toropi | 2* | 7 + 8 | 5 + 10 |
| Uri | Geuru/Ol | N | 7 + 8 | 2.2 + 12 |
| Younbaek | Keumkang/Tapdong | 2* | 13 + 16 | 2.2 + 12 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, Y.-R.; Beom, H.-R.; Altenbach, S.B.; Lee, M.-K.; Lim, S.-H.; Lee, J.-Y. Improved Method for Reliable HMW-GS Identification by RP-HPLC and SDS-PAGE in Common Wheat Cultivars. Molecules 2017, 22, 1055. https://doi.org/10.3390/molecules22071055
Jang Y-R, Beom H-R, Altenbach SB, Lee M-K, Lim S-H, Lee J-Y. Improved Method for Reliable HMW-GS Identification by RP-HPLC and SDS-PAGE in Common Wheat Cultivars. Molecules. 2017; 22(7):1055. https://doi.org/10.3390/molecules22071055
Chicago/Turabian StyleJang, You-Ran, Hye-Rang Beom, Susan B. Altenbach, Min-Ki Lee, Sun-Hyung Lim, and Jong-Yeol Lee. 2017. "Improved Method for Reliable HMW-GS Identification by RP-HPLC and SDS-PAGE in Common Wheat Cultivars" Molecules 22, no. 7: 1055. https://doi.org/10.3390/molecules22071055





