Mechanism Exploration of Arylpiperazine Derivatives Targeting the 5-HT2A Receptor by In Silico Methods
Abstract
:1. Introduction
2. Materials and Methods
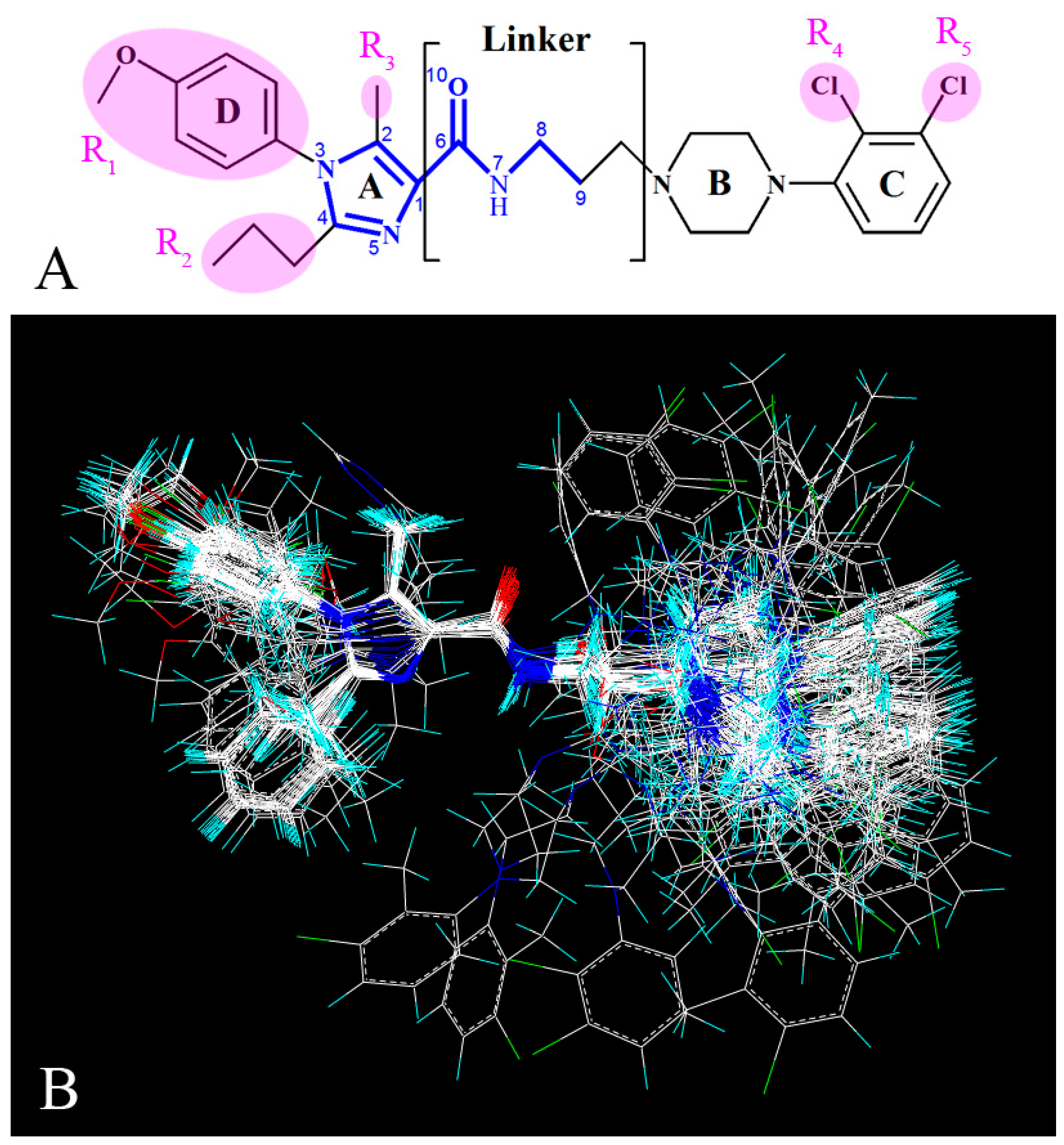
2.1. Data Set and Biological Activities
2.2. Molecular Modeling and Alignment
2.3. CoMFA and CoMSIA Studies
2.4. Molecular Docking
2.5. Molecular Dynamics
3. Results
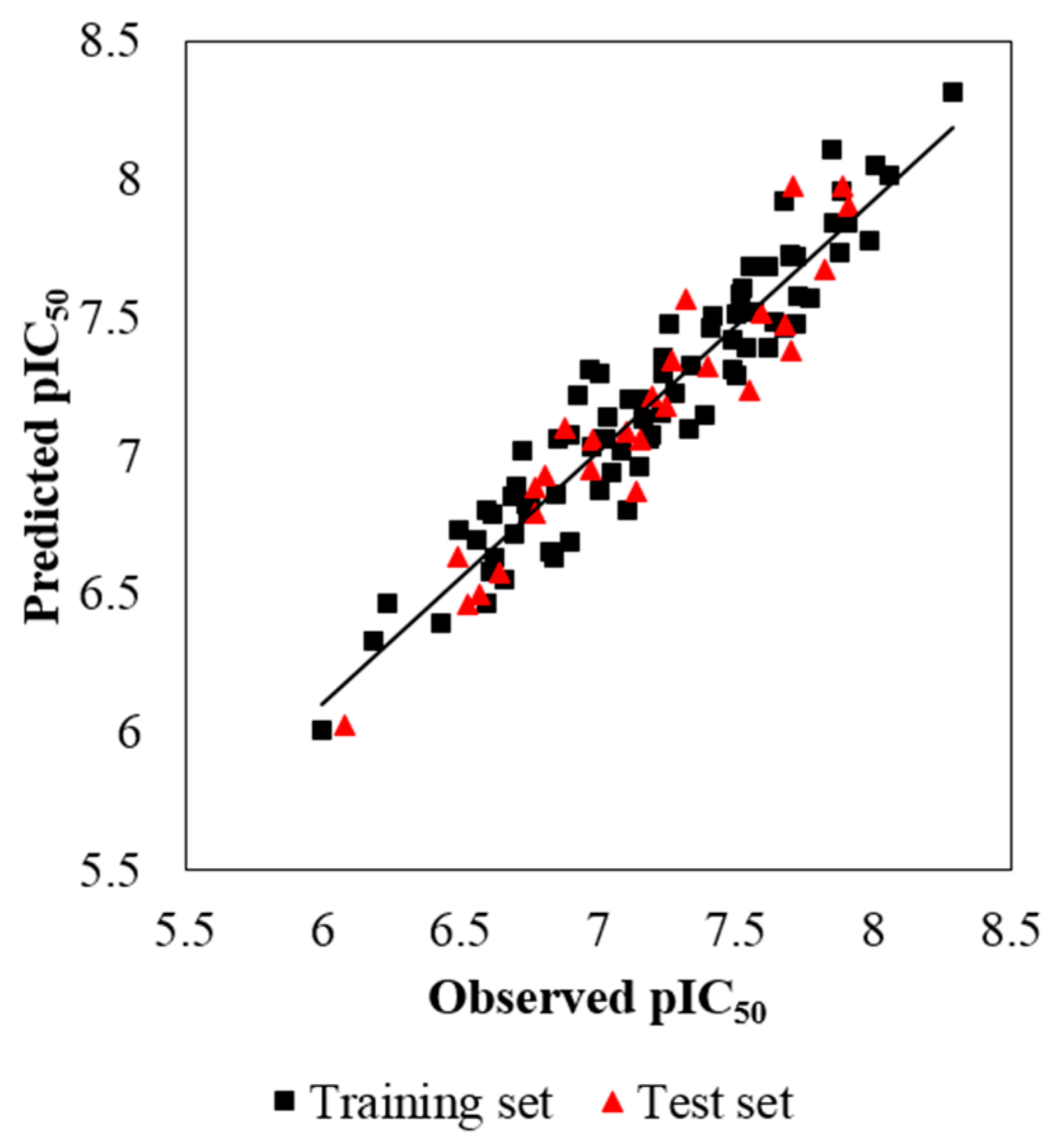
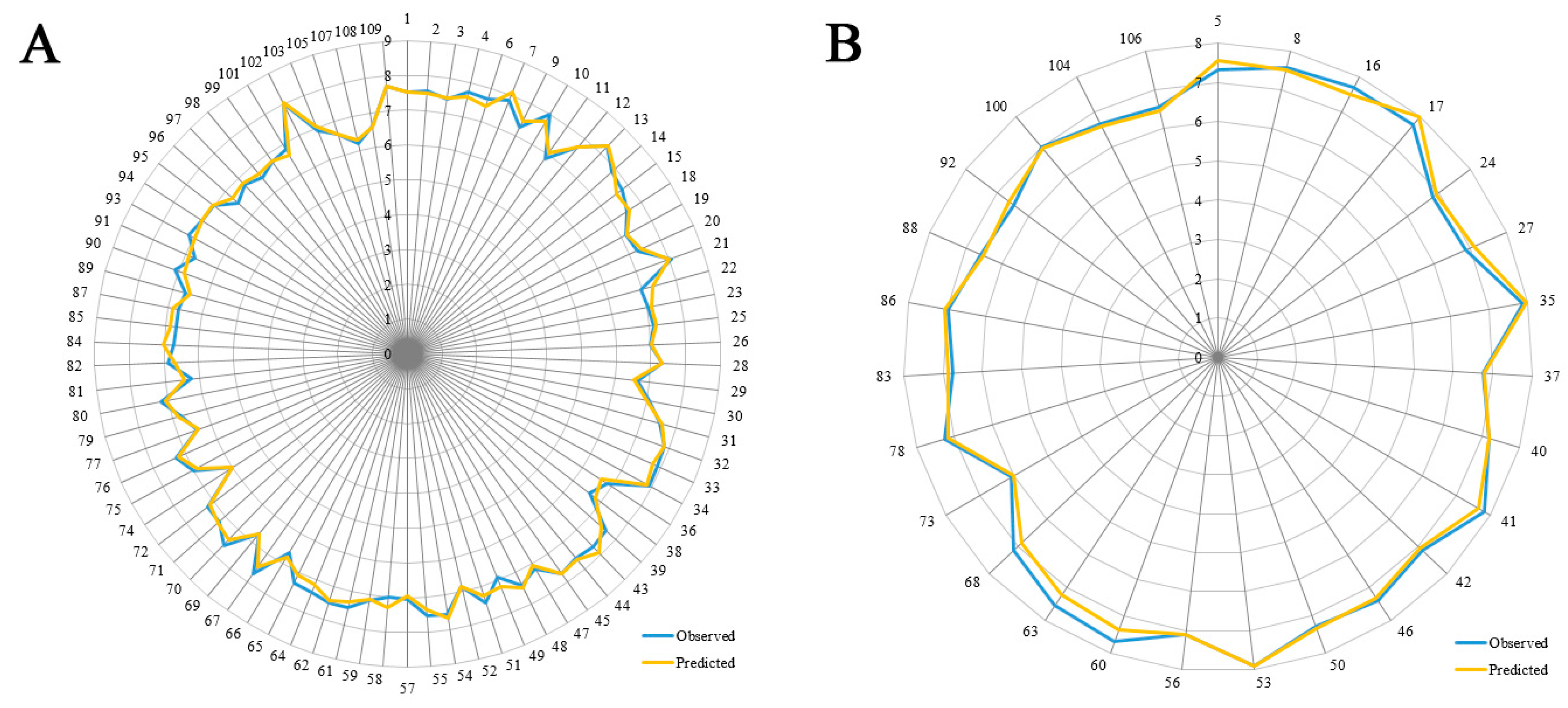
3.1. 3D-QSAR Statistical Analysis
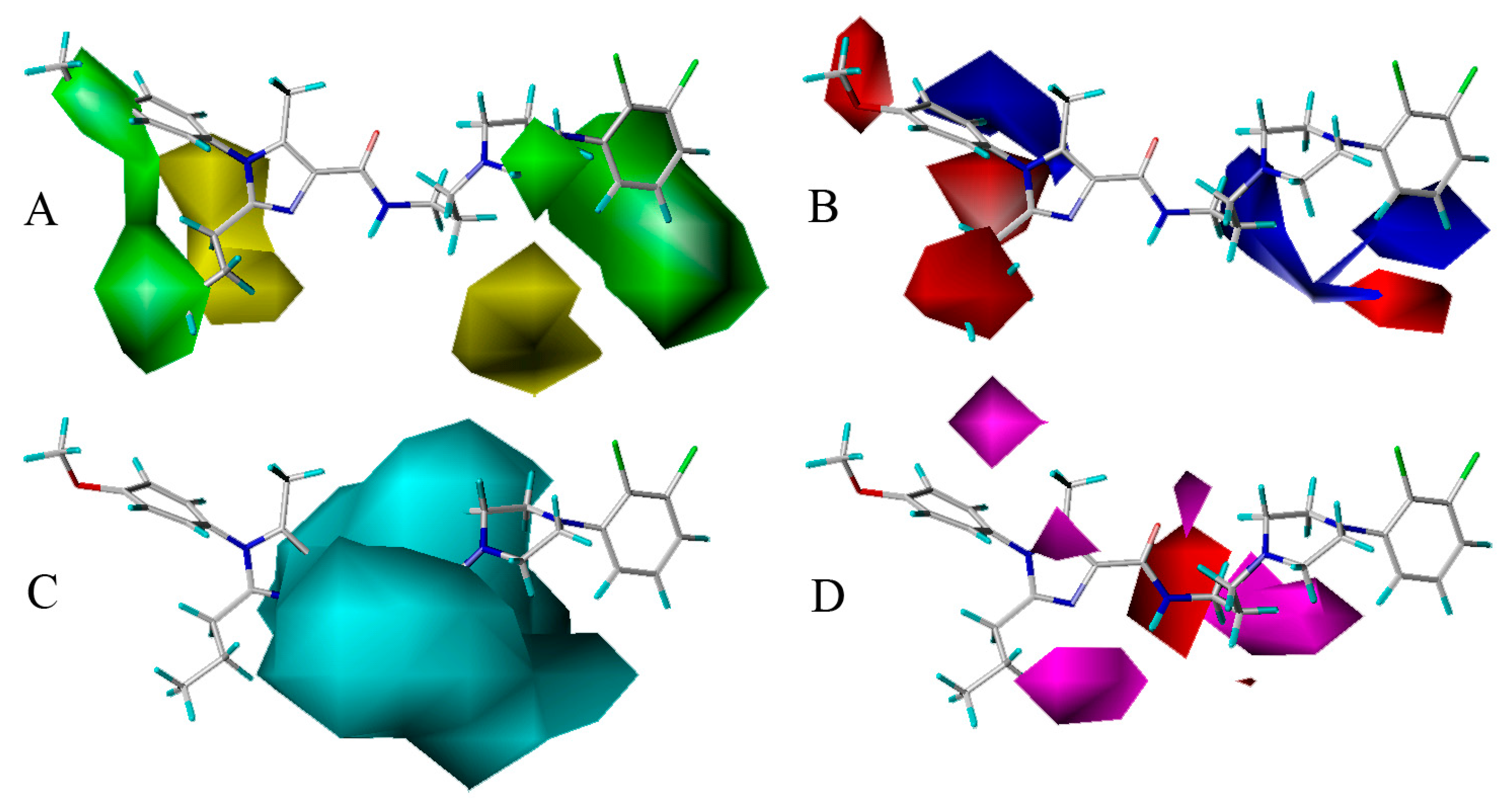
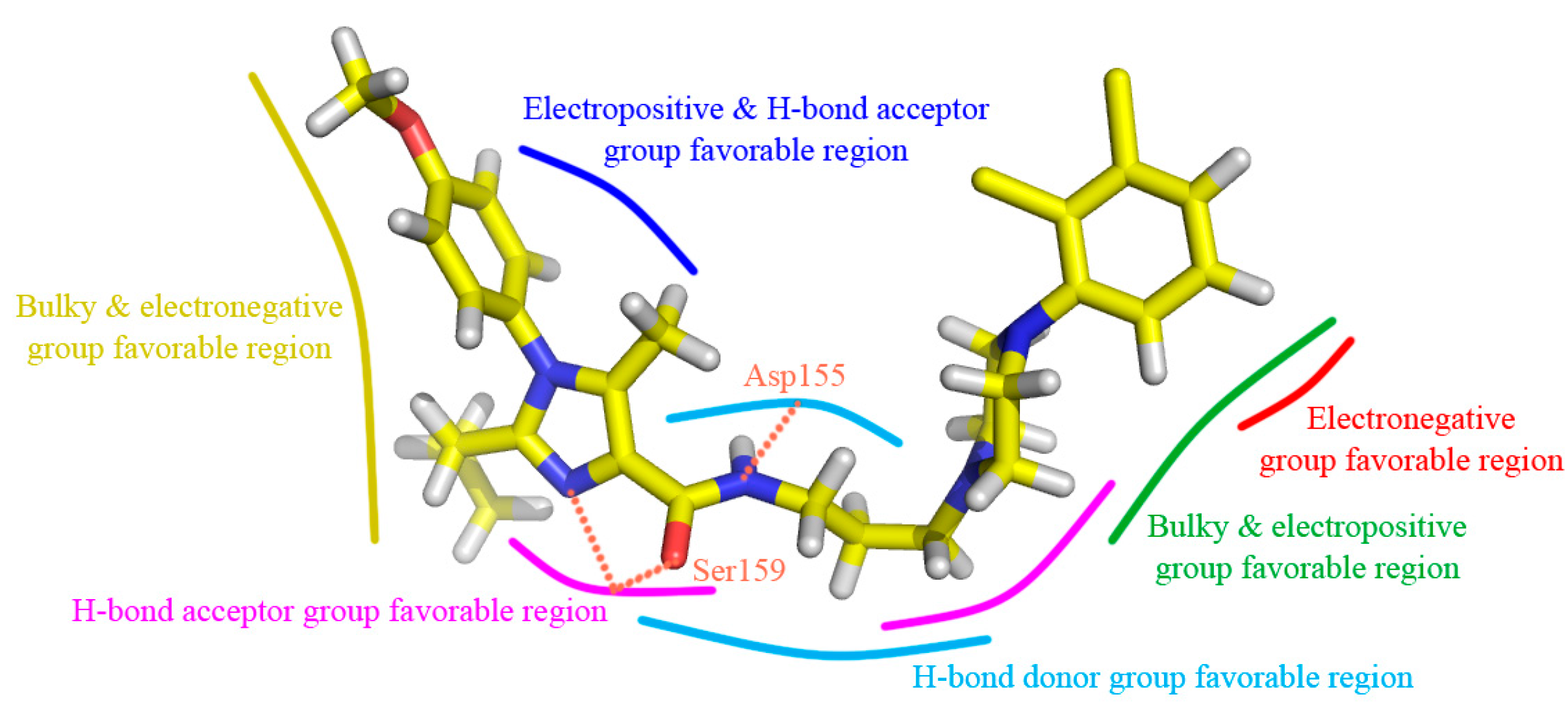
3.2. CoMSIA Graphical Interpretation
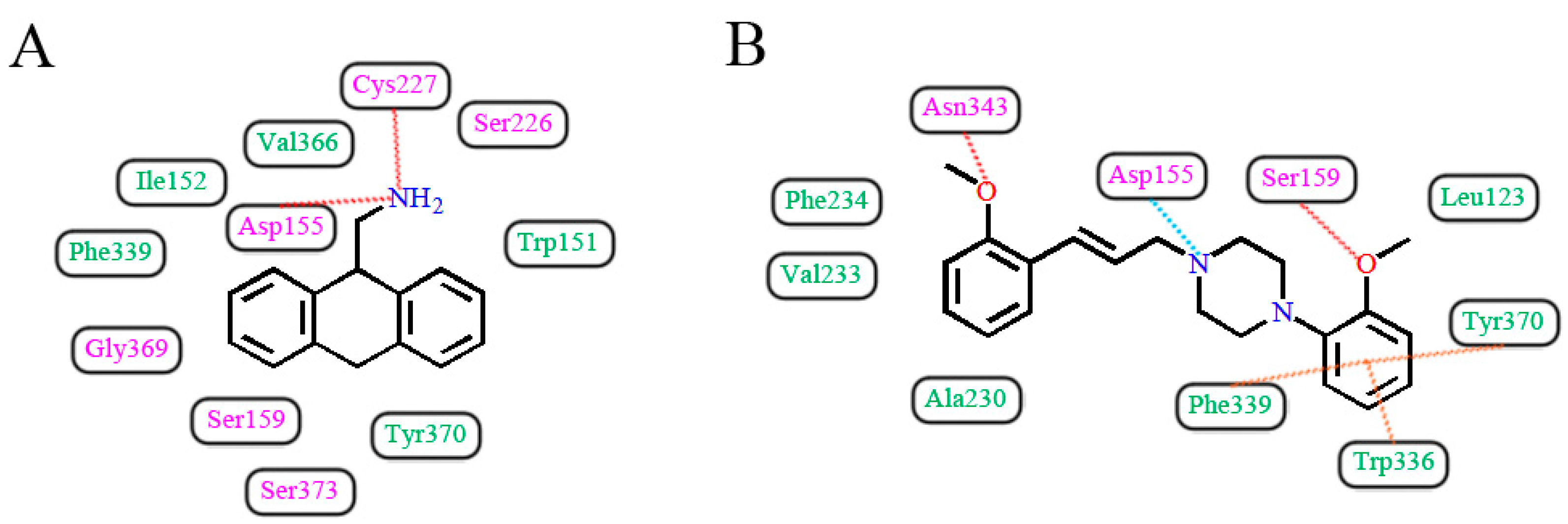
3.3. Docking Results
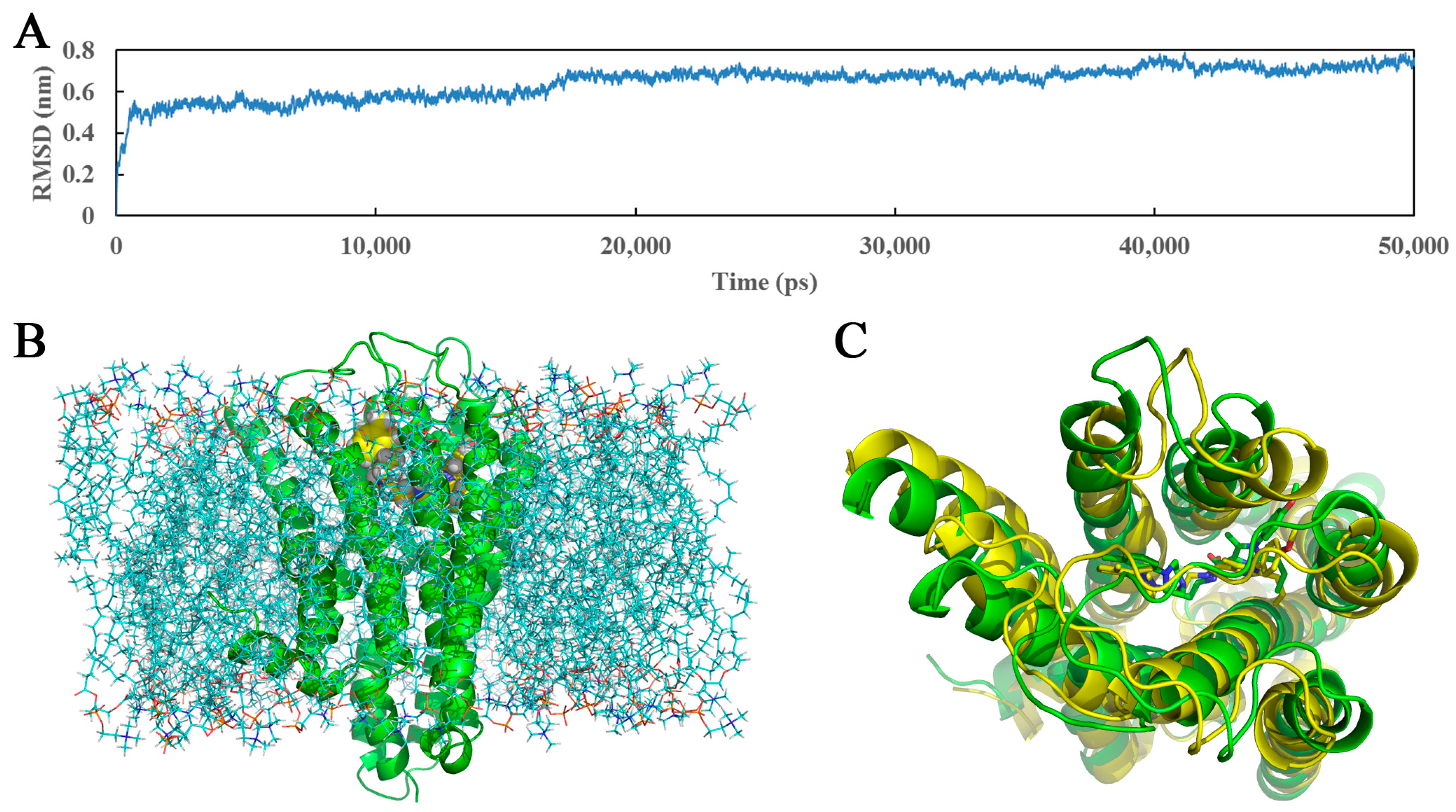
3.4. Molecular Dynamics Studies
3.5. Docking Comparison
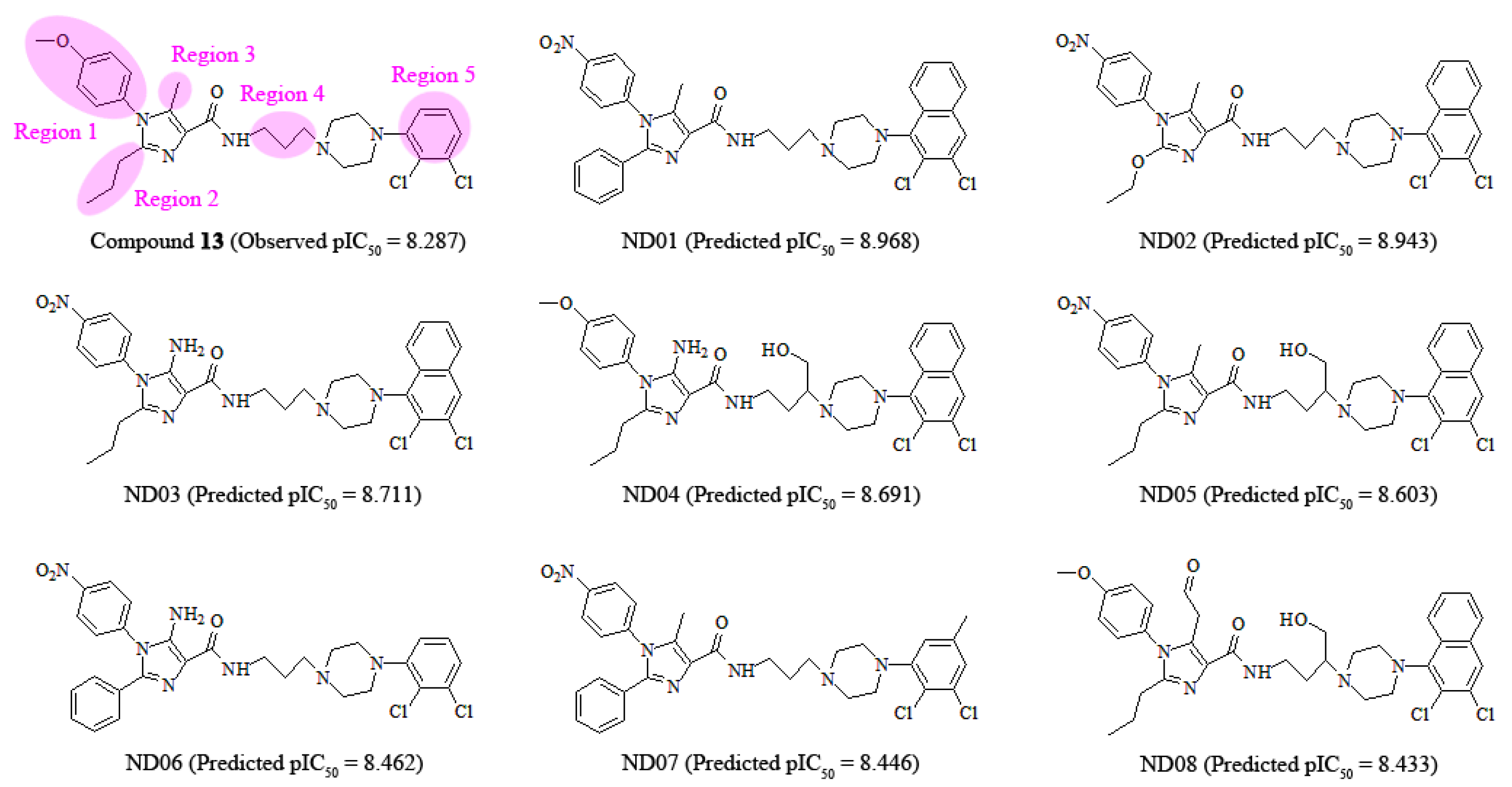
3.6. Design of New 5-HT2A Antagonists
4. Conclusions
- (1)
- The optimal CoMSIA model exhibits a statistically predictable ability with Q2 = 0.587, R2ncv = 0.900 and R2pre = 0.897, proving its wonderful reliability and predictability.
- (2)
- Bulky groups at R1 position and ring C, electropositive substituents at R3 and ring B, electronegative groups at R1, R2 positions and ring C, H-bond donor substituents at linker chain, H-bond acceptor groups at 10-position, ring A and ring B are favorable to increase the inhibitory activity.
- (3)
- The scaffold of antagonists fits into the conventional Site 2 of 5-HT2A receptor with an approximately “V” conformation and follows the second binding mode, which is fixed by three H-bonds with Asp155 and Ser159, two π-π stacking interactions with Try151 and Phe340, and hydrophobic interaction.
- (4)
- Several new potential arylpiperazine antagonists of 5-HT2A receptor were also newly-designed based on these results.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boulougouris, V.; Glennon, J.C.; Robbins, T.W. Dissociable effects of selective 5-HT2A and 5-HT2C receptor antagonists on serial spatial reversal learning in rats. Neuropsychopharmacology 2008, 33, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Poyurovsky, M.; Koren, D.; Gonopolsky, I.; Schneidman, M.; Fuchs, C.; Weizman, A.; Weizman, R. Effect of the 5-HT2 antagonist mianserin on cognitive dysfunction in chronic schizophrenia patients: An add-on, double-blind placebo-controlled study. Eur. Neuropsychopharm. 2003, 13, 123–128. [Google Scholar] [CrossRef]
- Seo, H.J.; Park, E.; Kim, M.J.; Kang, S.Y.; Lee, S.H.; Kim, H.J.; Lee, K.N.; Jung, M.E.; Lee, M.; Kim, M.; et al. Design and Synthesis of Novel Arylpiperazine Derivatives Containing the Imidazole Core Targeting 5-HT2A Receptor and 5-HT Transporter. J. Med. Chem. 2011, 54, 6305–6318. [Google Scholar] [CrossRef] [PubMed]
- Caspi, A.; Sugden, K.; Moffitt, T.E.; Taylor, A.; Craig, I.W.; Harrington, H.; McClay, J.; Mill, J.; Martin, J.; Braithwaite, A.; et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science 2003, 301, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J.; Barrot, M.; DiLeone, R.J.; Eisch, A.J.; Gold, S.J.; Monteggia, L.M. Neurobiology of Depression. Neuron 2002, 34, 13–25. [Google Scholar] [CrossRef]
- Shajib, M.S.; Khan, W.I. The role of serotonin and its receptors in activation of immune responses and inflammation. Acta Physiol. 2015, 213, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Carli, M.; Baviera, M.; Invernizzi, R.W.; Balducci, C. Dissociable contribution of 5-HT1A and 5-HT2A receptors in the medial prefrontal cortex to different aspects of executive control such as impulsivity and compulsive perseveration in rats. Neuropsychopharmacology 2006, 31, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Akhondzadeh, S.; Malek-Hosseini, M.; Ghoreishi, A.; Raznahan, M.; Rezazadeh, S. Effect of ritanserin, a 5HT2A/2C antagonist, on negative symptoms of schizophrenia: A double-blind randomized placebo-controlled study. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2008, 32, 1879–1883. [Google Scholar] [CrossRef] [PubMed]
- Talvik-Lotfi, M.; Nyberg, S.; Nordstrom, A.L.; Ito, H.; Halldin, C.; Brunner, F.; Farde, L. High 5HT2A receptor occupancy in M100907-treated schizophrenic patients. Psychopharmacology (Berl.) 2000, 148, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Umbricht, D.; Vollenweider, F.X.; Schmid, L.; Grubel, C.; Skrabo, A.; Huber, T.; Koller, R. Effects of the 5-HT2A agonist psilocybin on mismatch negativity generation and AX-continuous performance task: Implications for the neuropharmacology of cognitive deficits in schizophrenia. Neuropsychopharmacology 2003, 28, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Barr, A.M.; Lehmann-Masten, V.; Paulus, M.; Gainetdinov, R.R.; Caron, M.G.; Geyer, M.A. The selective serotonin-2A receptor antagonist M100907 reverses behavioral deficits in dopamine transporter knockout mice. Neuropsychopharmacology 2004, 29, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Vanover, K.E.; Davis, R.E. Role of 5-HT2A receptor antagonists in the treatment of insomnia. Nat. Sci. Sleep 2010, 2, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S.T.; Blier, P. Effects of serotonin (5-hydroxytryptamine, 5-HT) reuptake inhibition plus 5-HT2A receptor antagonism on the firing activity of norepinephrine neurons. J. Pharmacol. Exp. Ther. 2002, 302, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Pullar, I.A.; Carney, S.L.; Colvin, E.M.; Lucaites, V.L.; Nelson, D.L.; Wedley, S. LY367265, an inhibitor of the 5-hydroxytryptamine transporter and 5-hydroxytryptamine(2A) receptor antagonist: A comparison with the antidepressant, nefazodone. Eur. J. Pharmacol. 2000, 407, 39–46. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, D.; Kang, S.Y.; Park, W.; Kim, H.J.; Jung, M.E.; Son, E.; Pae, A.N.; Kim, J.; Lee, J. Arylpiperazine-containing pyrimidine 4-carboxamide derivatives targeting serotonin 5-HT2A, 5-HT2C, and the serotonin transporter as a potential antidepressant. Bioorg. Med. Chem. Lett. 2010, 20, 6439–6442. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Han, L.; Ren, Y. In Silico Exploration of 1,7-Diazacarbazole Analogs as Checkpoint Kinase 1 Inhibitors by Using 3D QSAR, Molecular Docking Study, and Molecular Dynamics Simulations. Molecules 2016, 21, 591. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Fu, X.; Wang, C.; Jiang, S.; Wang, J.; Zhang, S.; Yang, L.; Li, Y. QSAR, Molecular Docking and Molecular Dynamics of 3C-like Protease Inhibitors. Acta Phys. Chim. Sin. 2016, 32, 2693–2708. [Google Scholar]
- Li, Y.; Wang, J.; Xiao, Y.; Wang, Y.; Chen, S.; Yang, Y.; Lu, A.; Zhang, S. A systems pharmacology approach to investigate the mechanisms of action of Semen Strychni and Tripterygium wilfordii Hook F for treatment of rheumatoid arthritis. J. Ethnopharmacol. 2015, 175, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Li, Y.; Wang, Y. Computational Study Exploring the Interaction Mechanism of Benzimidazole Derivatives as Potent Cattle Bovine Viral Diarrhea Virus Inhibitors. J. Agric. Food Chem. 2016, 64, 5941–5950. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Yang, Y.; Zhang, J.; Du, J.; Zhang, S.; Yang, L. Profiling the interaction mechanism of indole-based derivatives targeting the HIV-1 gp120 receptor. RSC Adv. 2015, 5, 78278–78298. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Y.; Pan, Y.; Wang, J.; Lin, F.; Wang, C.; Zhang, S.; Yang, L. Computational Analysis of Structure-Based Interactions for Novel H1-Antihistamines. Int. J. Mol. Sci. 2016, 17, 129. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, J.; Li, Y.; Xiao, W.; Wang, Z.; Zhang, J.; Gao, W.; Zhang, S.; Yang, L. Structure determinants of indolin-2-on-3-spirothiazolidinones as MptpB inhibitors: An in silico study. Soft Matter 2013, 9, 11054. [Google Scholar] [CrossRef]
- Wu, M.; Li, Y.; Fu, X.; Wang, J.; Zhang, S.; Yang, L. Profiling the Interaction Mechanism of Quinoline/Quinazoline Derivatives as MCHR1 Antagonists: An in Silico Method. Int. J. Mol. Sci. 2014, 15, 15475–15502. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Klebe, G.; Abraham, U.; Mietzner, T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J. Med. Chem. 1994, 37, 4130–4146. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Yang, Y.; Zhang, S.; Yang, L. Profiling the Structural Determinants of Heteroarylnitrile Scaffold-Based Derivatives as Falcipain-2 Inhibitors by In Silico Methods. Curr. Med. Chem. 2013, 20, 2032–2042. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; van Opdenbosch, N. Validation of the general purpose Tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Wang, Q.; Mach, R.H.; Reichert, D.E. Docking and 3D-QSAR Studies on Isatin Sulfonamide Analogues as Caspase-3 Inhibitors. J. Chem. Inf. Model. 2009, 49, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lei, M.; Lu, A.; Zhao, X.; Yin, X.; Gao, Q. 3D-QSAR studies of boron-containing dipeptides as proteasome inhibitors with CoMFA and CoMSIA methods. Eur. J. Med. Chem. 2009, 44, 1486–1499. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, S.; Jaiswal, S.; Katti, S.B.; Prabhakar, Y.S. CoMFA and CoMSIA analysis of tetrahydroquinolines as potential antimalarial agents. SAR QSAR Environ. Res. 2011, 22, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Sjöström, M.; Eriksson, L. Partial Least Squares Projections to Latent Structures (PLS) in Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2002. [Google Scholar]
- Sprous, D.G.; Palmer, R.K.; Swanson, J.T.; Lawless, M. QSAR in the Pharmaceutical Research Setting: QSAR Models for Broad, Large Problems. Curr. Top. Med. Chem. 2010, 10, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Kovalishyn, V.V.; Kholodovych, V.; Tetko, I.V.; Welsh, W.J. Volume learning algorithm significantly improved PLS model for predicting the estrogenic activity of xenoestrogens. J. Mol. Graph. Model. 2007, 26, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Yang, L.; Zhang, S.; Liu, C.; Yang, S. Comparison of steroid substrates and inhibitors of P-glycoprotein by 3D-QSAR analysis. J. Mol. Struct. 2005, 733, 111–118. [Google Scholar] [CrossRef]
- Thaimattam, R.; Daga, P.; Rajjak, S.A.; Banerjee, R.; Iqbal, J. 3D-QSAR CoMFA, CoMSIA studies on substituted ureas as Raf-1 kinase inhibitors and its confirmation with structure-based studies. Bioorgan. Med. Chem. 2004, 12, 6415–6425. [Google Scholar] [CrossRef] [PubMed]
- Shahlaei, M.; Madadkar-Sobhani, A.; Mahnam, K.; Fassihi, A.; Saghaie, L.; Mansourian, M. Homology modeling of human CCR5 and analysis of its binding properties through molecular docking and molecular dynamics simulation. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 802–817. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Westkaemper, R.B.; Runyon, S.P.; Savage, J.E.; Roth, B.L.; Glennon, R.A. Exploring the relationship between binding modes of 9-(aminomethyl)-9,10-dihydroanthracene and cyproheptadine analogues at the 5-HT2A serotonin receptor. Bioorg. Med. Chem. Lett. 2001, 11, 563–566. [Google Scholar] [CrossRef]
- Runyon, S.P.; Mosier, P.D.; Roth, B.L.; Glennon, R.A.; Westkaemper, R.B. Potential Modes of Interaction of 9-Aminomethyl-9,10-dihydroanthracene (AMDA) Derivatives with the 5-HT2A Receptor: A Ligand Structure-Affinity Relationship, Receptor Mutagenesis and Receptor Modeling Investigation. J. Med. Chem. 2008, 51, 6808–6828. [Google Scholar] [CrossRef] [PubMed]
- Kanagarajadurai, K.; Malini, M.; Bhattacharya, A.; Panicker, M.M.; Sowdhamini, R. Molecular modeling and docking studies of human 5-hydroxytryptamine 2A (5-HT2A) receptor for the identification of hotspots for ligand binding. Mol. Biosyst. 2009, 5, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Yap, B.K.; Buckle, M.J.C.; Doughty, S.W. Homology modeling of the human 5-HT1A, 5-HT2A, D1, and D2 receptors: Model refinement with molecular dynamics simulations and docking evaluation. J. Mol. Model. 2012, 18, 3639–3655. [Google Scholar] [CrossRef] [PubMed]
- Sencanski, M.; Sukalovic, V.; Shakib, K.; Soskic, V.; Dosen-Micovic, L.; Kostic-Rajacic, S. Molecular Modeling of 5HT(2A) Receptor—Arylpiperazine Ligands Interactions. Chem. Biol. Drug Des. 2014, 83, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Ponnala, S.; Kapadia, N.; Navarro, H.A.; Harding, W.W. Aporphinoid Antagonists of 5-HT2A Receptors: Further Evaluation of Ring A Substituents and the Size of Ring C. Chem. Biol. Drug Des. 2014, 84, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Guo, L.; Xu, L.; Zhen, X.; Yu, K.; Zhao, W.; Fu, W. Discovery of novel potent and selective ligands for 5-HT2A receptor with quinazoline scaffold. Bioorg. Med. Chem. Lett. 2015, 25, 3970–3974. [Google Scholar] [CrossRef] [PubMed]
- Gandhimathi, A.; Sowdhamini, R. Molecular modelling of human 5-hydroxytryptamine receptor (5-HT2A) and virtual screening studies towards the identification of agonist and antagonist molecules. J. Biomol. Struct. Dyn. 2016, 34, 952–970. [Google Scholar] [CrossRef] [PubMed]
- Ísberg, V.; Balle, T.; Sander, T.; Jørgensen, F.S.; Gloriam, D.E. G Protein- and Agonist-Bound Serotonin 5-HT2A Receptor Model Activated by Steered Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011, 51, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- McRobb, F.M.; Capuano, B.; Crosby, I.T.; Chalmers, D.K.; Yuriev, E. Homology Modeling and Docking Evaluation of Aminergic G Protein-Coupled Receptors. J. Chem. Inf. Model. 2010, 50, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Cordova-Sintjago, T.; Sakhuja, R.; Kondabolu, K.; Canal, C.E.; Booth, R.G. Molecular Determinants for Ligand Binding at Serotonin 5-HT2A and 5-HT2C GPCRs: Experimental Affinity Results Analyzed by Molecular Modeling and Ligand Docking Studies. Int. J. Quantum Chem. 2012, 112, 3807–3814. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Im, W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Galeazzi, R.; Massaccesi, L.; Piva, F.; Principato, G.; Laudadio, E. Insights into the influence of 5-HT2c aminoacidic variants with the inhibitory action of serotonin inverse agonists and antagonists. J. Mol. Model. 2014, 20, 2120. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Perryman, A.L.; Schames, J.R.; McCammon, J.A. Computational Drug Design Accommodating Receptor Flexibility: The Relaxed Complex Scheme. J. Am. Chem. Soc. 2002, 124, 5632–5633. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.; Fraaije, J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Wu, Q.; Li, X.; Gao, Q.; Wang, J.; Li, Y.; Yang, L. Interaction mechanism exploration of HEA derivatives as BACE1 inhibitors by in silico analysis. Mol. Biosyst. 2016, 12, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, C.; Wang, J.; Yang, Y.; Zhang, J.; Zhang, S.; Yang, L. Insight into the Structural Features of Pyrazolopyrimidine- and Pyrazolopyridine-based B-RafV600E Kinase Inhibitors by Computational Explorations. Chem. Biol. Drug Des. 2014, 83, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Yang, Y.; Du, J.; Zhang, S.; Yang, L. In silico research to assist the investigation of carboxamide derivatives as potent TRPV1 antagonists. Mol. Biosyst. 2015, 11, 2885–2899. [Google Scholar] [CrossRef] [PubMed]
- Van Vlijmen, H.W.T.; Schaefer, M.; Karplus, M. Improving the accuracy of protein pKa calculations: Conformational averaging versus the average structure. Proteins Struct. Funct. Bioinform. 1998, 33, 145–158. [Google Scholar] [CrossRef]
- Westkaemper, R.B.; Glennon, R.A. Application of ligand SAR, receptor modeling and receptor mutagenesis to the discovery and development of a new class of 5-HT(2A) ligands. Curr. Top. Med. Chem. 2002, 2, 575–598. [Google Scholar] [CrossRef] [PubMed]
- Surgand, J.; Rodrigo, J.; Kellenberger, E.; Rognan, D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins Struct. Funct. Bioinform. 2006, 62, 509–538. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |





| No. | Structure | IC50 (nM) | No. | Structure | IC50 (nM) |
|---|---|---|---|---|---|
| 2 |  | 27 | 5 t |  | 48 |
| 13 |  | 5.17 | 18 |  | 30 |
| 32 |  | 13.9 | 46 t |  | 40 |
| 47 |  | 64.8 | 49 |  | 117.9 |
| 60 t |  | 20 | 74 |  | 1011 |
| 84 |  | 185 | 90 |  | 78.3 |
| 95 |  | 95 | 101 |  | 147 |
| 105 |  | 237 | 106 t |  | 294 |
| No. | Researchers | Template | Representative Structure | Site Type | Binding Interactions | Crucial Residues |
|---|---|---|---|---|---|---|
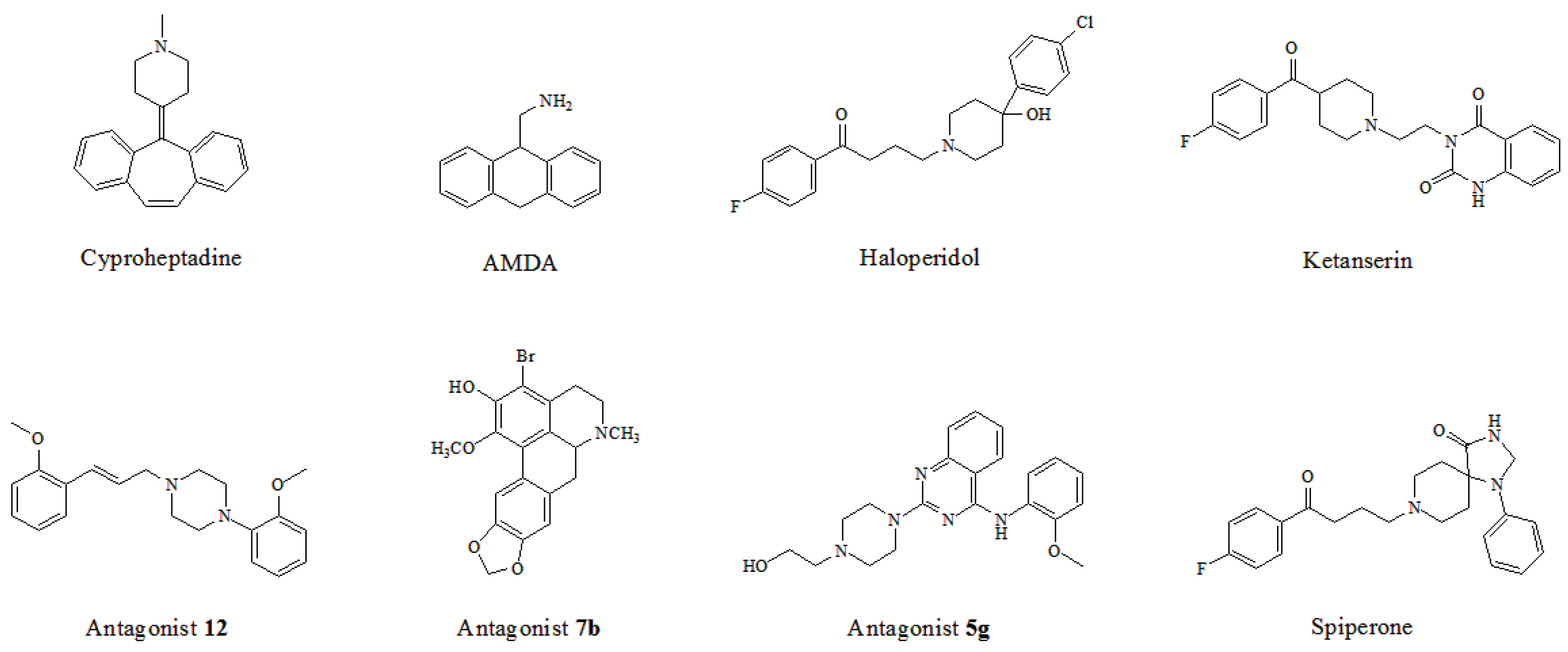
| 1 | Westkaemper et al. [39] | GF-62 cells | Cyproheptadine | Site 2 | H-bond | Asp155 |
| 2 | Runyon et al. [40] | Bovine rhodopsin (PDB entry 1U19:A) | AMDA | Site 2 | H-bond, hydrophobic interaction | Asp155, Cys227, Val366 |
| 3 | Kanagarajadurai et al. [41] | β2-adrenergic receptor (PDB entry 2RH1) | Haloperidol | Site 2 | H-bond, hydrophobic interaction | Asp155, Phe339, Phe340, Tyr370 |
| 4 | Yap et al. [42] | β2-adrenergic receptor (PDB entry 2RH1) | Ketanserin | Site 2 | H-bond | Thr134, Asp155, Phe234, Val235, Ser239 |
| 5 | Sencanski et al. [43] | β2-adrenergic receptor (PDB entry 3D4S) | Antagonist 12 | Site 2 | H-bond, π-π stacking, salt bridge | Asp155, Ser159, Trp336, Phe339, Asn343, Tyr370 |
| 6 | Ponnala et al. [44] | β2-adrenergic receptor (PDB entry 2RH1) | Antagonist 7b | Site 2 | H-bond, hydrophobic interaction | Asp155, Phe339, Phe340 |
| 7 | Deng et al. [45] | β2-adrenergic receptor | Antagonist 5g | Site 2 | H-bond, cation-π | Trp151, Asp155, Ser159, Tyr370 |
| 8 | Gandhimathi et al. [46] | β2-adrenergic receptor (PDB entry 2RH1) | Spiperone | Site 2 | H-bond, salt bridge | Asp155, Asn363 |
| 9 | Gandhimathi et al. [46] | β2-adrenergic receptor (PDB entry 3SN6) | Spiperone | Site 2 | H-bond, π-π stacking | Trp151, Asp155, Asn343 |
| PLS Statistics | CoMFA | CoMSIA |
| Q2 | 0.496 | 0.587 |
| R2ncv | 0.914 | 0.900 |
| R2pre | 0.831 | 0.897 |
| F | 132.803 | 94.879 |
| SEE | 0.148 | 0.161 |
| SEP | 0.402 | 0.339 |
| ONC | 6 | 7 |
| Field Contribution/% | ||
| Steric | 55.6 | 24.5 |
| Electrostatic | 44.4 | 45.4 |
| H-bond donor | - | 14.9 |
| H-bond acceptor | - | 15.2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Li, F.; Wang, C.; Wang, J.; Yang, Y.; Yang, L.; Li, Y. Mechanism Exploration of Arylpiperazine Derivatives Targeting the 5-HT2A Receptor by In Silico Methods. Molecules 2017, 22, 1064. https://doi.org/10.3390/molecules22071064
Lin F, Li F, Wang C, Wang J, Yang Y, Yang L, Li Y. Mechanism Exploration of Arylpiperazine Derivatives Targeting the 5-HT2A Receptor by In Silico Methods. Molecules. 2017; 22(7):1064. https://doi.org/10.3390/molecules22071064
Chicago/Turabian StyleLin, Feng, Feng Li, Chao Wang, Jinghui Wang, Yinfeng Yang, Ling Yang, and Yan Li. 2017. "Mechanism Exploration of Arylpiperazine Derivatives Targeting the 5-HT2A Receptor by In Silico Methods" Molecules 22, no. 7: 1064. https://doi.org/10.3390/molecules22071064




