Molecular Cloning and Characterization of PnbHLH1 Transcription Factor in Panax notoginseng
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cloning of PnbHLH1 Gene
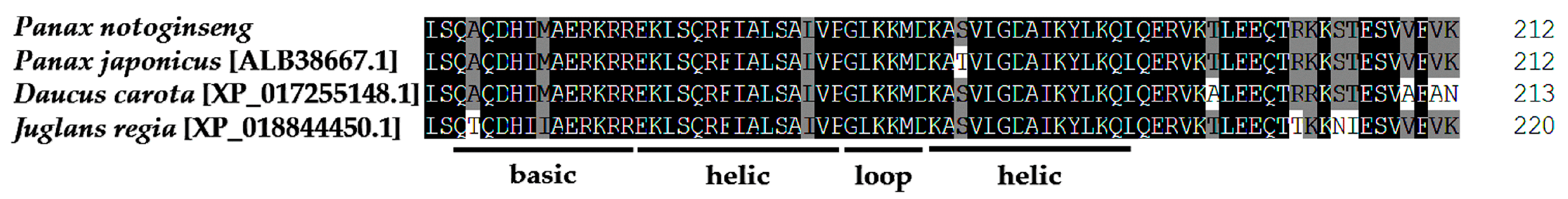
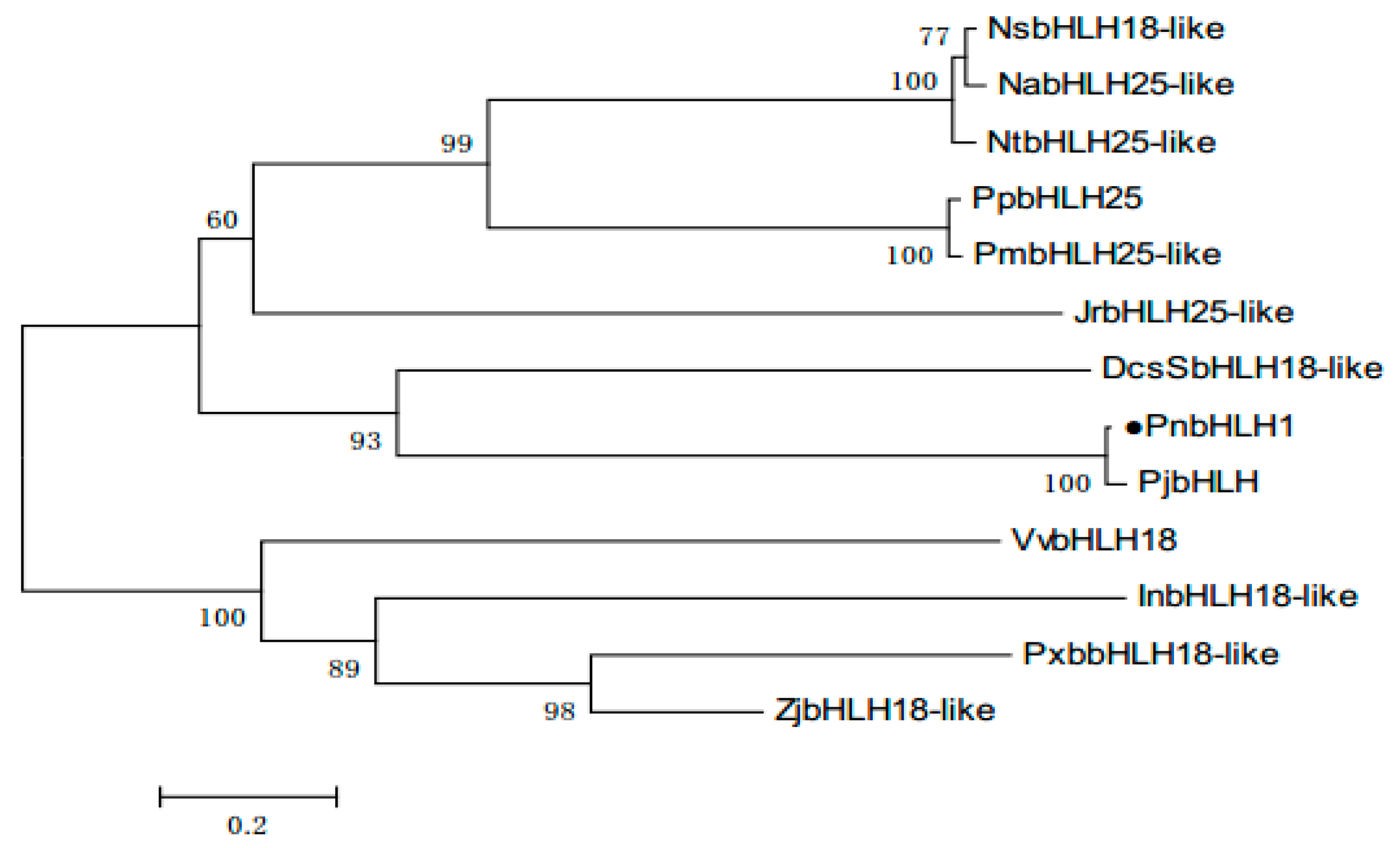
2.2. Bioinformatics Analysis of PnbHLH1 Transcription Factor
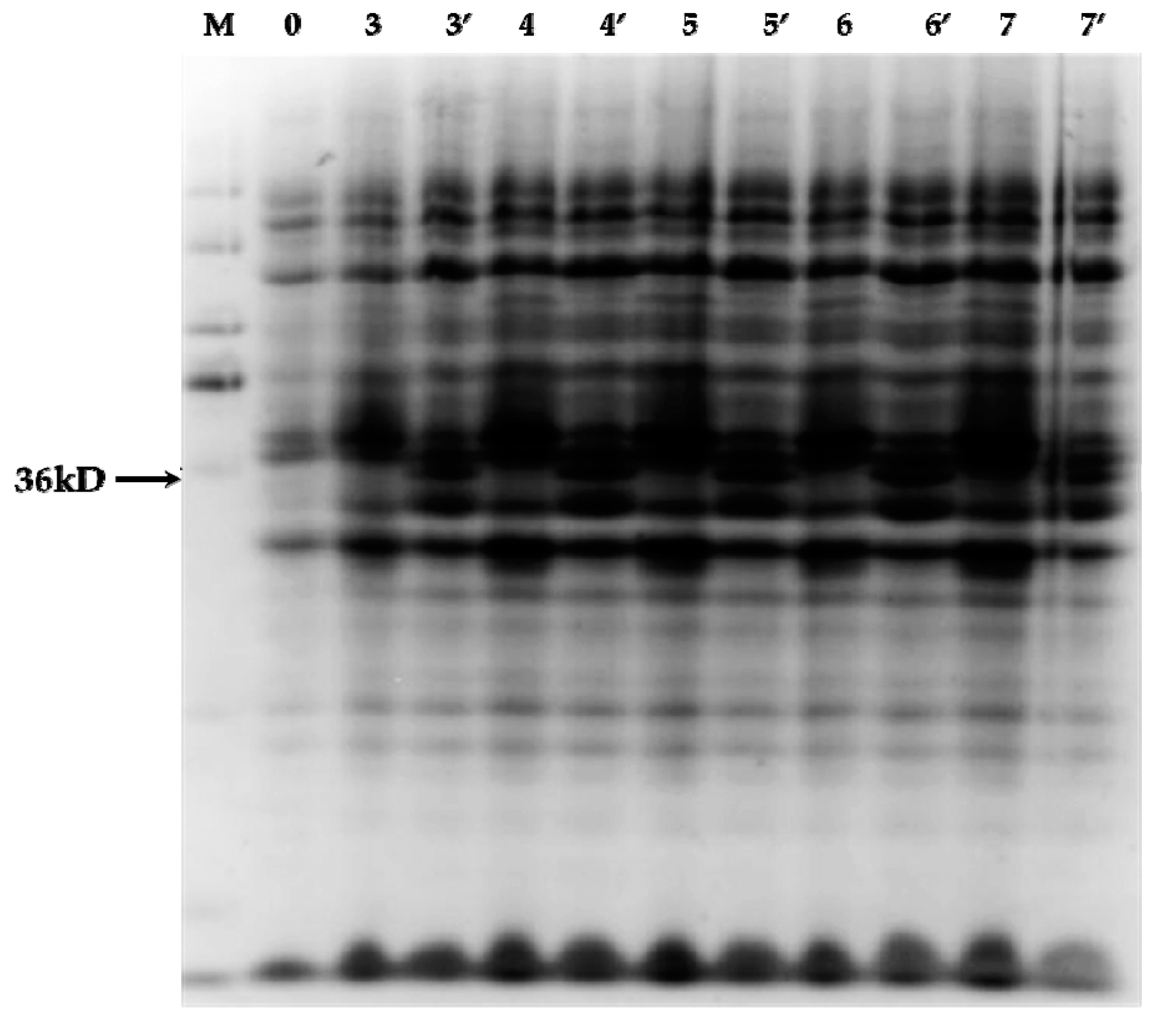
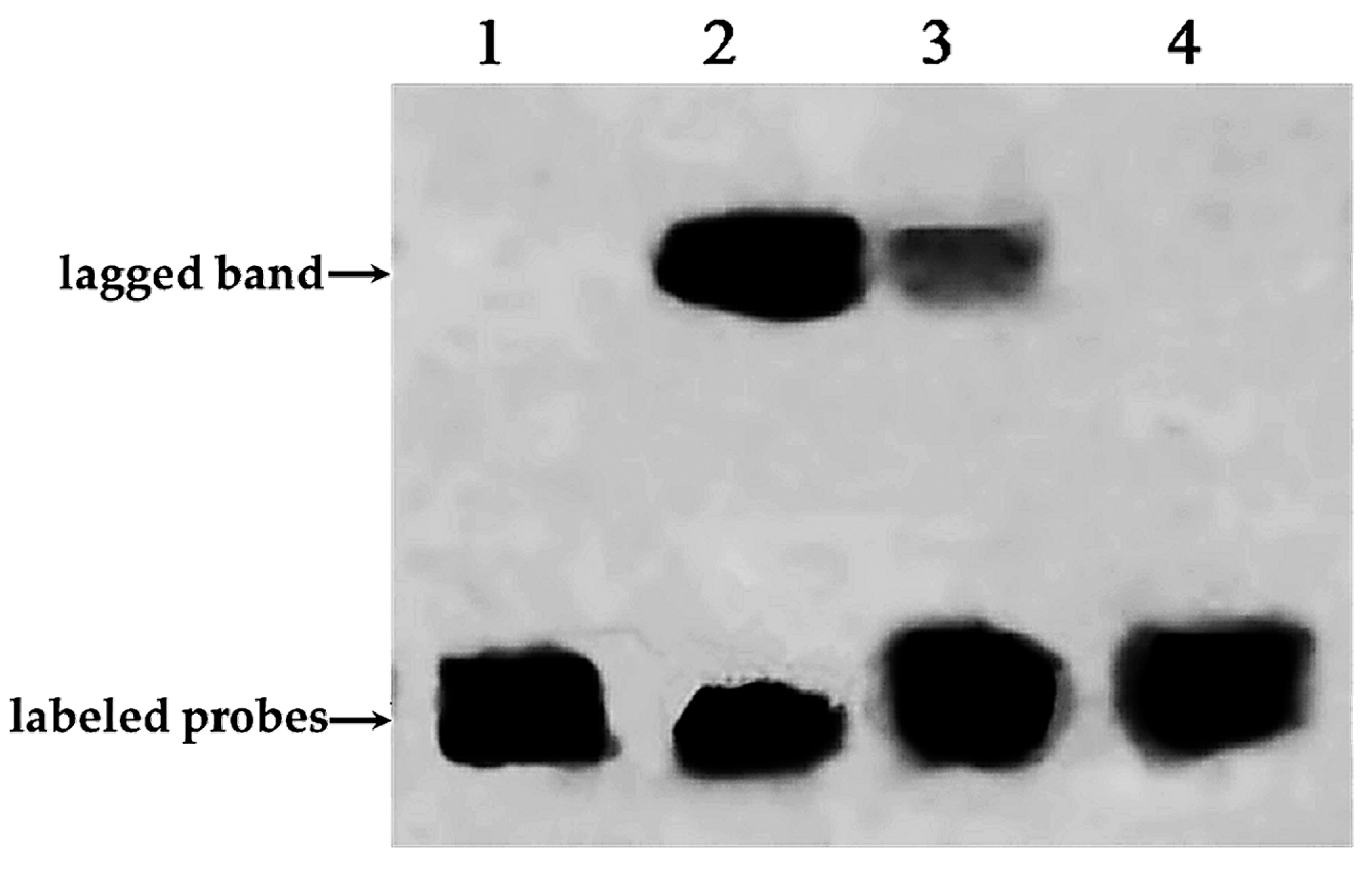
2.3. Electrophoretic Mobility Shift Assay Experiment of PnbHLH1 and Cis-Acting Element E-Box
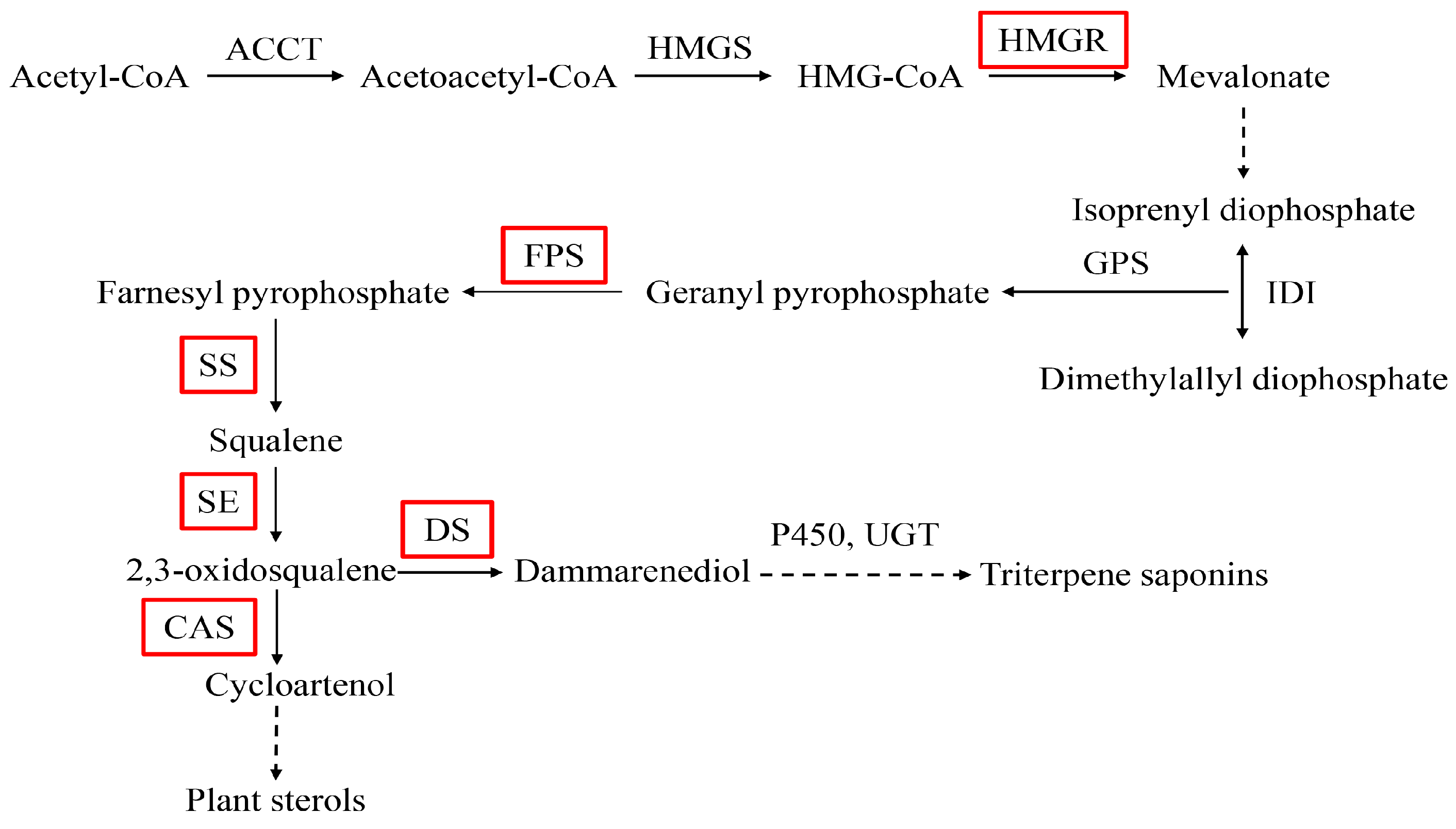
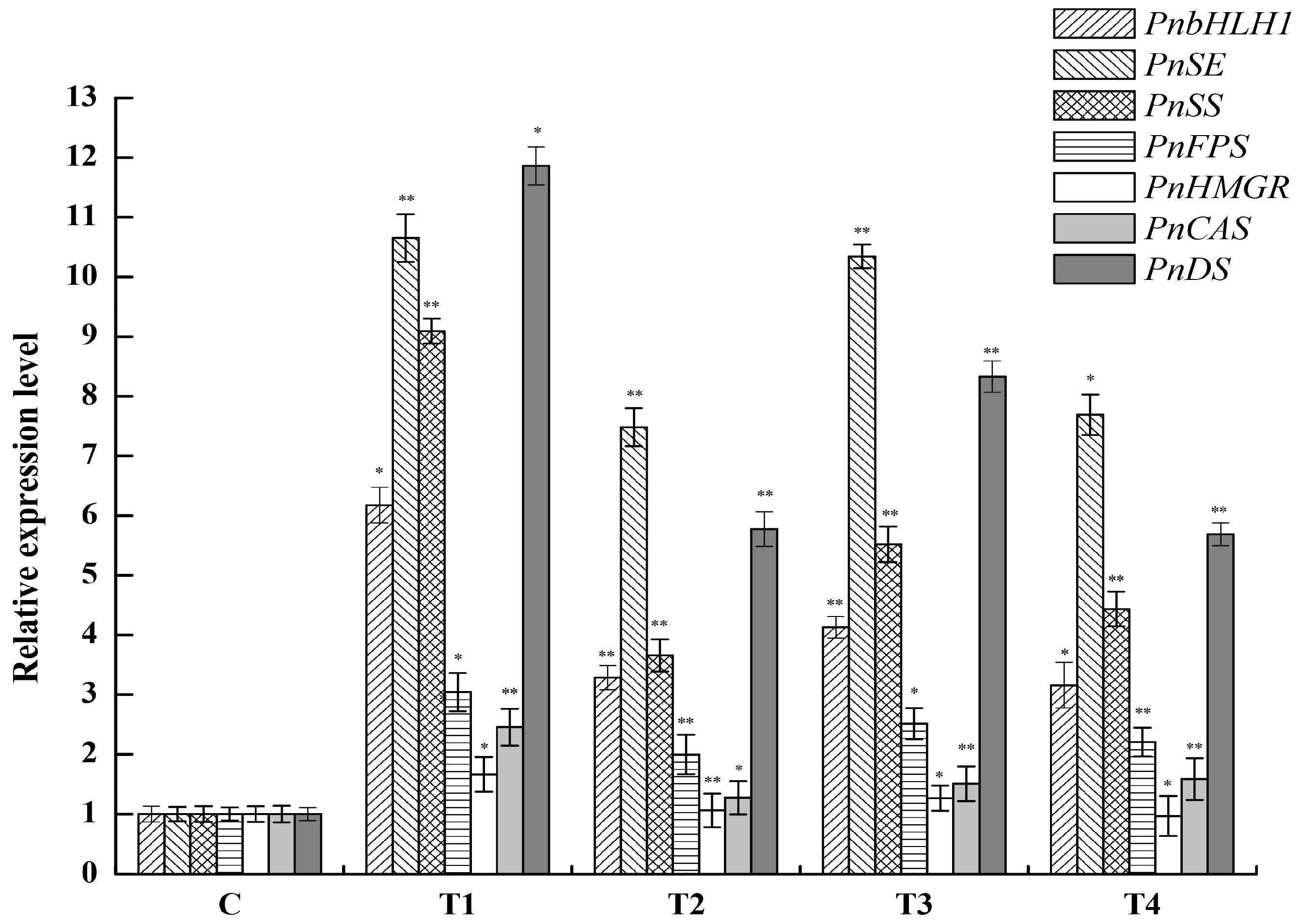
2.4. Expression Analysis of PnbHLH1 and Other Key Enzyme Genes in Transgenic Cells
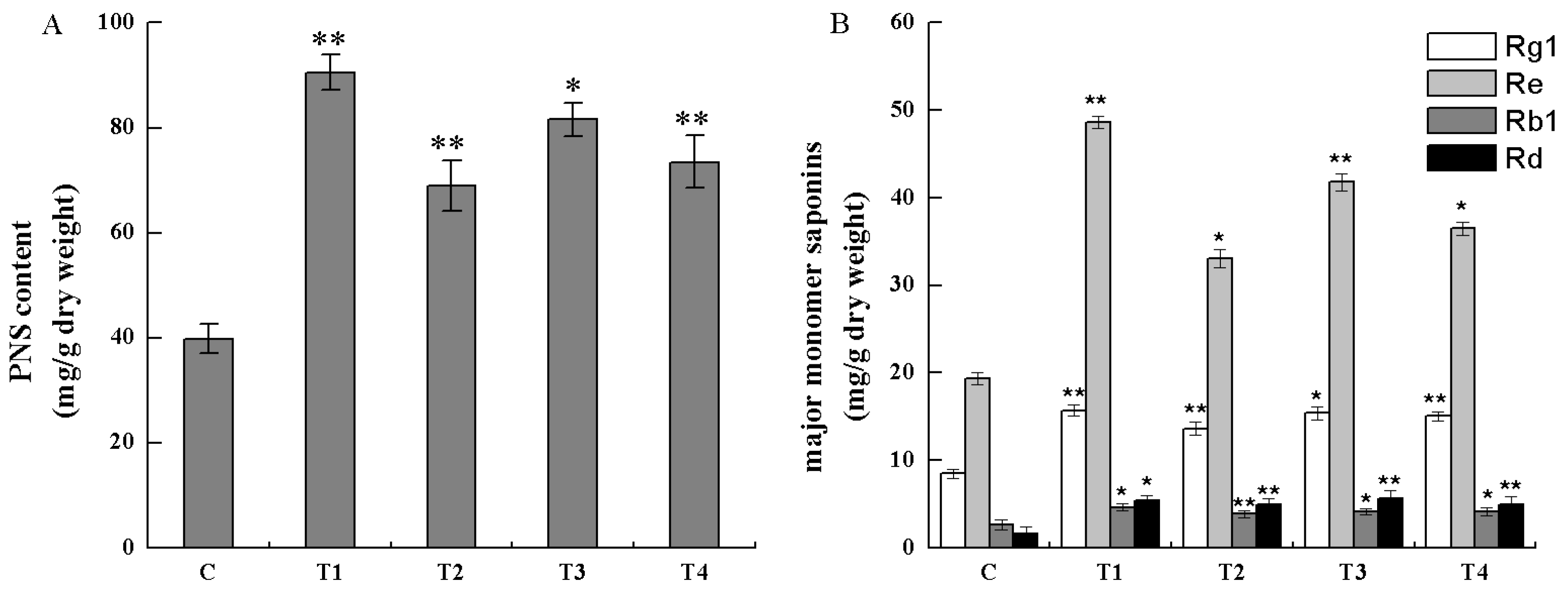
2.5 Analysis of Saponins in Transgenic Cells
3. Materials and Methods
3.1. Plant Materials
3.2. Gene Sequencing, Phylogenetic Gene Structure, and Conserved Motif Analyses
3.3. Constructions of Plant Expression Vector and Prokaryotic Expression Vector Carrying the PnbHLH1 Gene
3.4. Transformation of the PnbHLH1 Plant Expression Vector Mediated by Agrobacterium
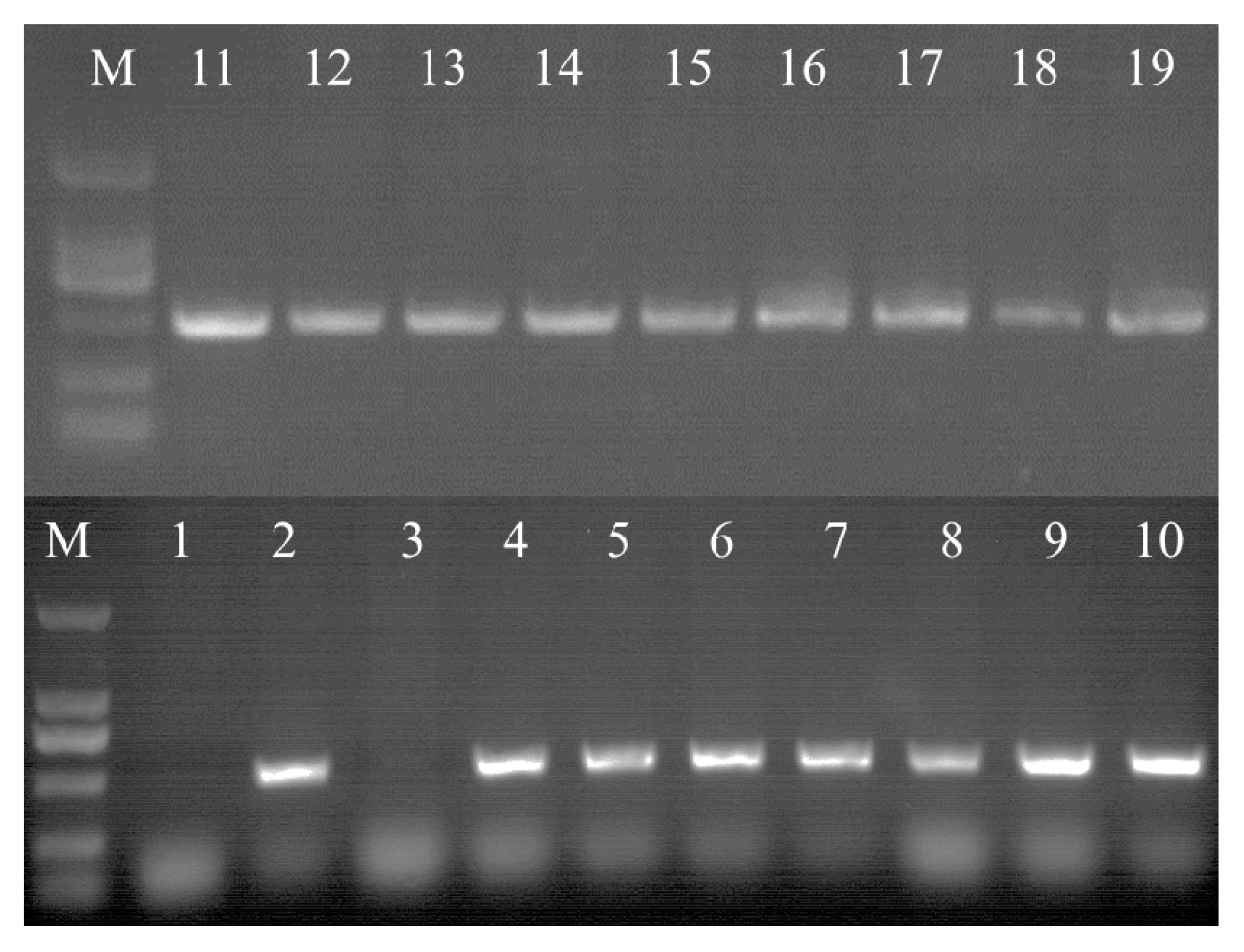
3.5. Identification of Transgenic Cells by PCR
3.6. Expression Analysis by qRT-PCR
3.7. Analysis of Triterpenoid Saponins
3.8. Analysis of PnbHLH1 Binding with DNA
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, T.; Guo, R.; Zhou, G.H.; Zhou, X.; Kou, Z.Z.; Sui, F.; Li, C.; Tang, L.Y.; Wang, Z.J. Traditional uses, botany, phytochemistry, pharmacology and toxicology of P. notoginseng (Burk.) F.H. Chen: A review. J. Ethnopharmacol. 2016, 188, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Kuzuyama, T. Mevalonate and Nonmevalonate Pathways for the Biosynthesis of Isoprene Units. Biosci. Biotechnol. Biochem. 2014, 66, 1619–1627. [Google Scholar] [CrossRef]
- Zhang, J.Z. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003, 6, 430–440. [Google Scholar] [CrossRef]
- Liu, L.S.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants-functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Atchley, W.R.; Fitch, W.M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C.Y.; Eachus, R.A.; Trieu, W.; Ro, D.K.; Keasling, J.D. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 2007, 3, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.L.; Qin, Q.L.; Dai, D.L.; Kong, L.S.; Li, W.; Zha, X.J.; Jin, Y.J.; Tang, K.X. Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata. Biotechnol. Lett. 2009, 31, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Z.; Niu, Y.Y.; Xu, J.; Li, Y.; Luo, H.M.; Zhu, Y.J.; Liu, M.Z.; Wu, Q.; Song, J.Y.; Sun, C.; et al. Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tissue Org. 2013, 114, 269–277. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, K.P.; Yao, Q. Progress of studies on bHLH transcription factor families. Hereditas 2008, 30, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.T.; Hedhili, S.; Montiel, G.; Zhang, Y.X.; Chatel, G.; Pre, M.; Gantet, P.; Memelink, J. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011, 67, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.T.; Ye, B.P. Progress in Study on Refolding and Renaturation of Inclusion Bodies. Pharm. Biotechnol. 2017, 14, 306–309. [Google Scholar]
- Gun, W.; Huang, L.; Xu, Z.; Cen, P. Purification and Refolding Escherichia coli Produced Enterokinase Inclusion Bodies Immobilized by Nickel Chelating Chromatography. J. Food Sci. Biotechnol. 2006, 25, 18–22. [Google Scholar]
- Ji, Y.P.; Xiao, J.W.; Shen, Y.L.; Ma, D.M.; Li, Z.Q.; Pu, G.B.; Li, X.; Huang, L.L.; Liu, B.Y.; Ye, H.C.; et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 55, 1592–1604. [Google Scholar] [CrossRef] [PubMed]
- Murthy, H.N.; Georgiev, M.I.; Kim, Y.S.; Jeong, C.S.; Kim, S.J.; Park, S.Y.; Paek, K.Y. Ginsenosides: Prospective for sustainable biotechnological production. Appl. Microbiol. Biotechnol. 2014, 98, 6243–6254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.P.; Wu, Y.Y.; Jin, J.; Hu, B.X.; Zeng, W.Y.; Zhu, W.J.; Zheng, Y.L.; Chen, P. De novo characterization of Panax japonicus C.A. Mey transcriptome and genes related to triterpenoid saponin biosynthesis. Biochem. Biophys. Res. Commun. 2015, 466, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Huang, Z.J.; Yu, H.; Wang, Y.; Liu, D.Q. Transformation of Panax notoginseng saponins by steaming and Trichoderma longibrachiatum. Biotechnol. Biotechnol. Equip. 2016, 30, 165–172. [Google Scholar] [CrossRef]
- Yang, Y.; Ge, F.; Sun, Y.; Liu, D.Q.; Chen, C.Y. Strengthening triterpene saponins biosynthesis by over-expression of farnesyl pyrophosphate synthase gene and RNA interference of cycloartenol synthase gene in Panax notoginseng cells. Molecules 2017, 22, 581. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.W.; Ge, F.; Sun, Y.; Liu, D.Q.; Chen, C.Y. Transcription factors involved in plant terpenoid biosynthesis and their application prospect. Chin. Tradit. Herb. Drugs 2012, 43, 2512–2519. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Hofgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef] [PubMed]
- Puissant, C.; Houdebine, L.M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques 1990, 8, 148–149. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Zhang, P.; Ge, F.; Liu, D.Q.; Chen, C.Y. Enhancement of triterpenoid saponins biosynthesis in Panax notoginseng cells by co-overexpressions of 3-hydroxy-3-methylglutaryl CoA reductase and squalene synthase genes. Biochem. Eng. J. 2017, 122, 38–46. [Google Scholar] [CrossRef]
Sample Availability: Samples are not available from the authors. |


| Primer Name | Primer Sequence (5′–3′) |
|---|---|
| HptⅡ-F | GAAGTGCTTGACATTGGGGAAT |
| HptⅡ-R | AGATGTTGGCGACCTCGTATT |
| 18S rRNA-F | AACCATAAACGATGCCGACCAG |
| 18S rRNA-R | TTCAGCCTTGCGACCATACTCC |
| PnSS-F | GCAGGACTTGTTGGATTAGGGT |
| PnSS-R | AACATGCGTGACTTTGGTATCTC |
| PnHMGR-F | GGCAGGACCCAGCACAAAATA |
| PnHMGR-R | ACACCCAGAAGGTTCAAGCAA |
| PnFPS-F | CGGATGCTGGACTATAATGTG |
| PnFPS-R | ATTTACGGCAATCATACCAACC |
| PnDS-F | TTTGGGAGCCTCCAGTTCCAAAACC |
| PnDS-R | TCTCTTTCTGACGATGCCCAGGATG |
| PnSE-F | TCTCCATGACTCATCAACCCTC |
| PnSE-R | CGGCCAACGCCATAGATAG |
| PnCAS-F | CGGATGGTTTATCTGCCTATGTC |
| PnCAS-R | GGTTGCGTGCTTGATTCCA |
| PnbHLH1-F | ACCTATTGATCTTATGACCTCCCA |
| PnbHLH1-R | CATCTTCTTTAGGCCAGGGA |
| Primer | Primer Sequence (5′–3′) |
|---|---|
| E-box-F | AATGCANNTGTTGGGGAGCANNTGTTGGGGAGCANNTGGGGA |
| E-box-R | TCCCCANNTGCTCCCCAACANNTGCTCCCCAACANNTGCATT |
| mE-box-F | AATGATTATTTTGGGGAGATTATTTTGGGGAGATTATTGGGA |
| mE-box-R | TCCCAATAATCTCCCCAAAATAATCTCCCCAAAATAATCATT |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ge, F.; Deng, B.; Shah, T.; Huang, Z.; Liu, D.; Chen, C. Molecular Cloning and Characterization of PnbHLH1 Transcription Factor in Panax notoginseng. Molecules 2017, 22, 1268. https://doi.org/10.3390/molecules22081268
Zhang X, Ge F, Deng B, Shah T, Huang Z, Liu D, Chen C. Molecular Cloning and Characterization of PnbHLH1 Transcription Factor in Panax notoginseng. Molecules. 2017; 22(8):1268. https://doi.org/10.3390/molecules22081268
Chicago/Turabian StyleZhang, Xiang, Feng Ge, Bing Deng, Taif Shah, Zhuangjia Huang, Diqiu Liu, and Chaoyin Chen. 2017. "Molecular Cloning and Characterization of PnbHLH1 Transcription Factor in Panax notoginseng" Molecules 22, no. 8: 1268. https://doi.org/10.3390/molecules22081268




