Transcriptional Responses of Creeping Bentgrass to 2,3-Butanediol, a Bacterial Volatile Compound (BVC) Analogue
Abstract
:1. Introduction
2. Results
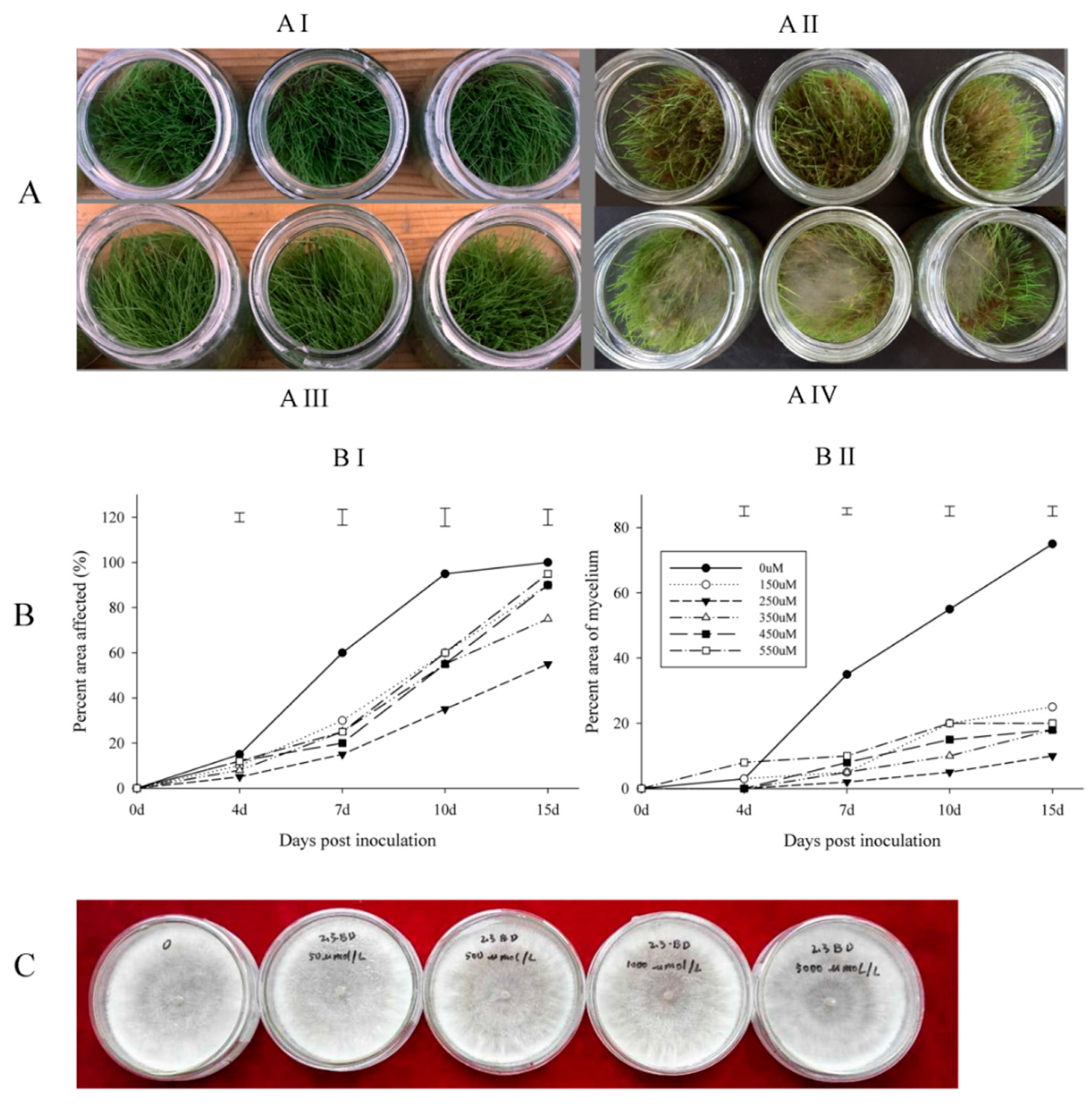
2.1. Disease Assessment of Creeping Bentgrass Treated with BD and the Antimicrobial Activity of BD
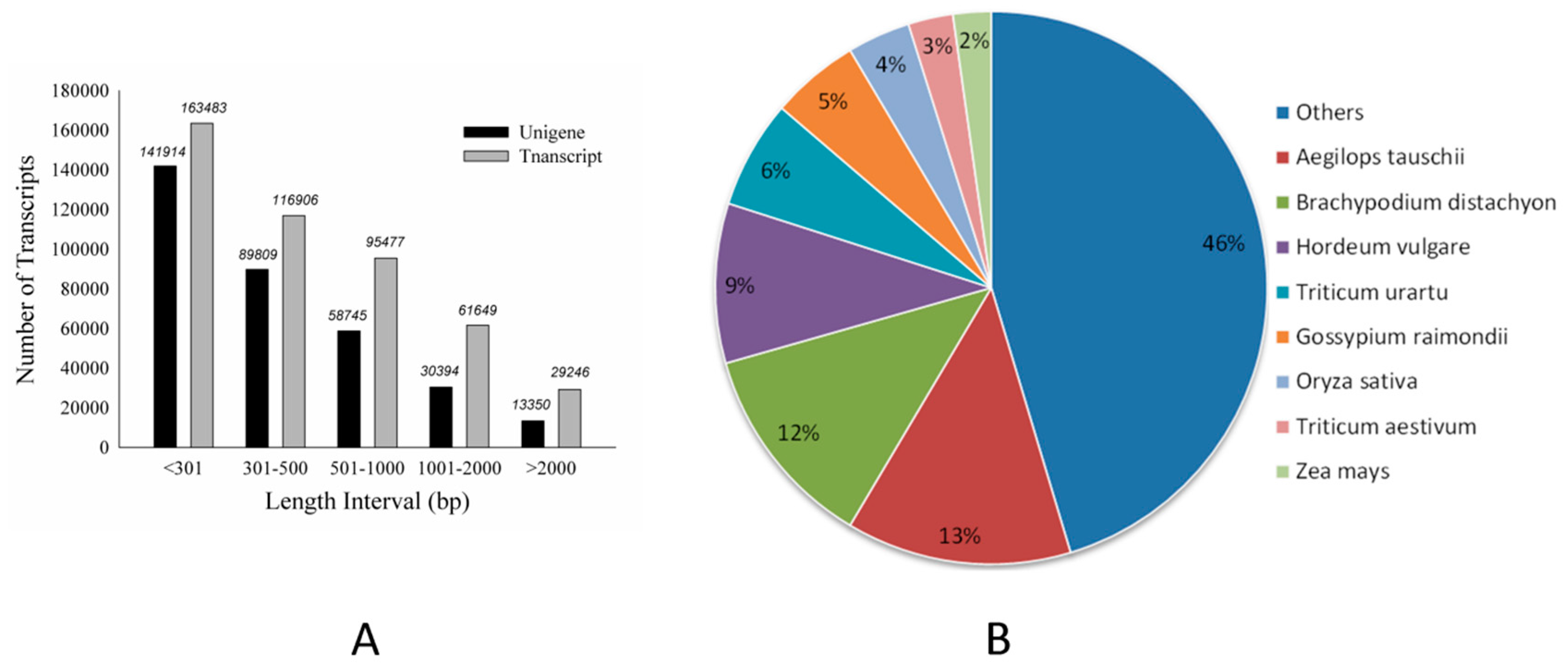
2.2. Transcriptome Sequencing and Read Statistics
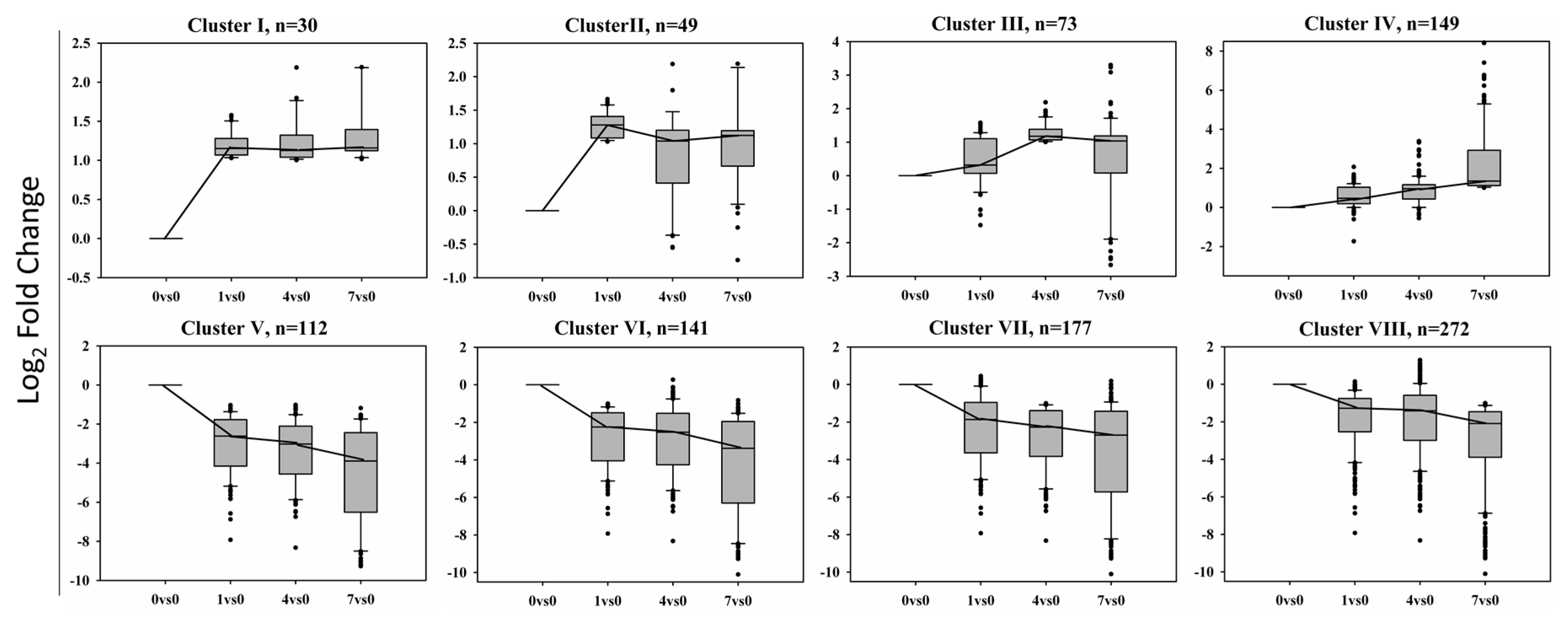
2.3. Differentially Expressed Genes under the Treatment of BD
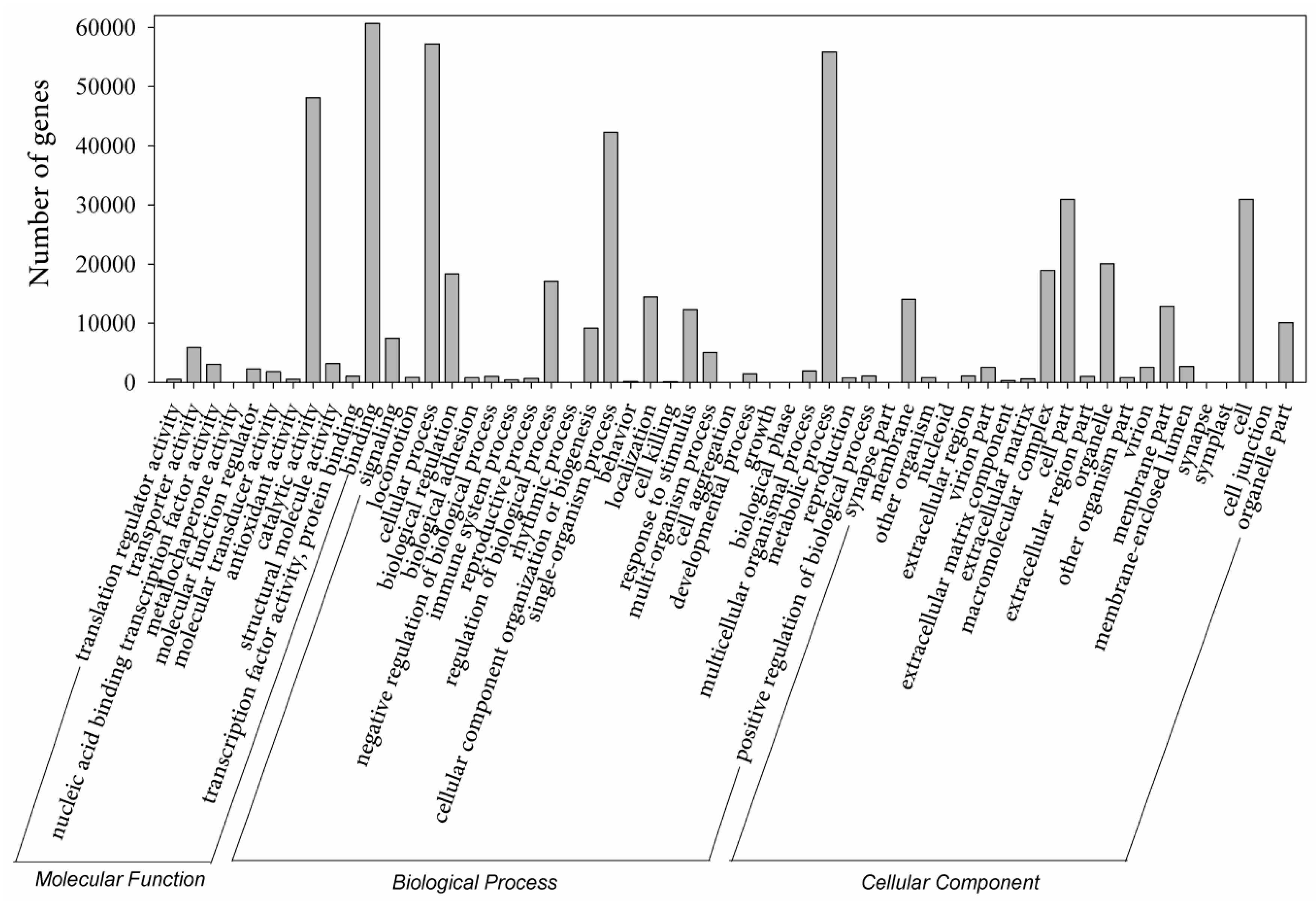
2.4. Gene Ontology Enrichment Analysis of Eight Clusters
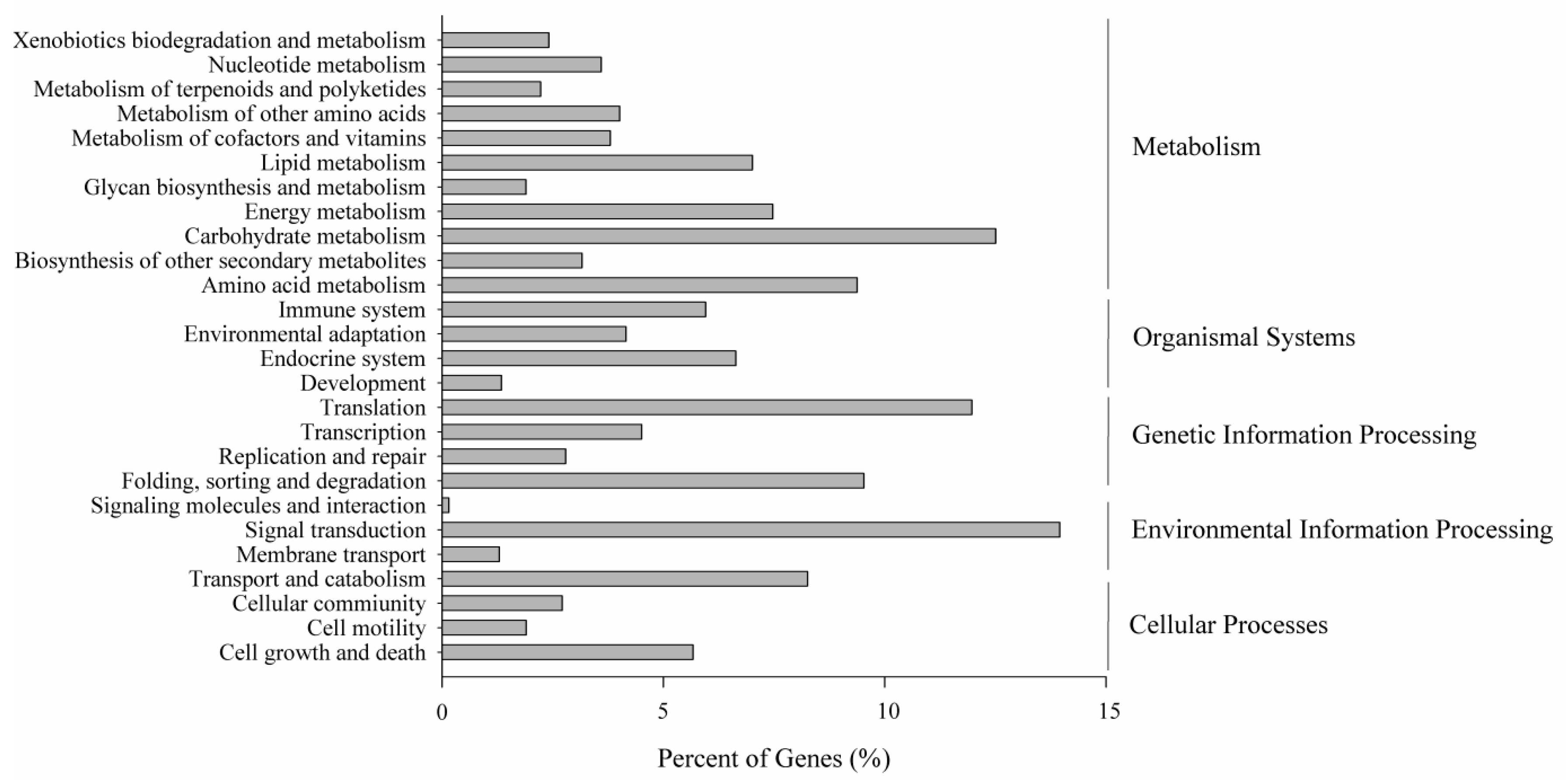
2.5. Metabolic Pathway Analysis
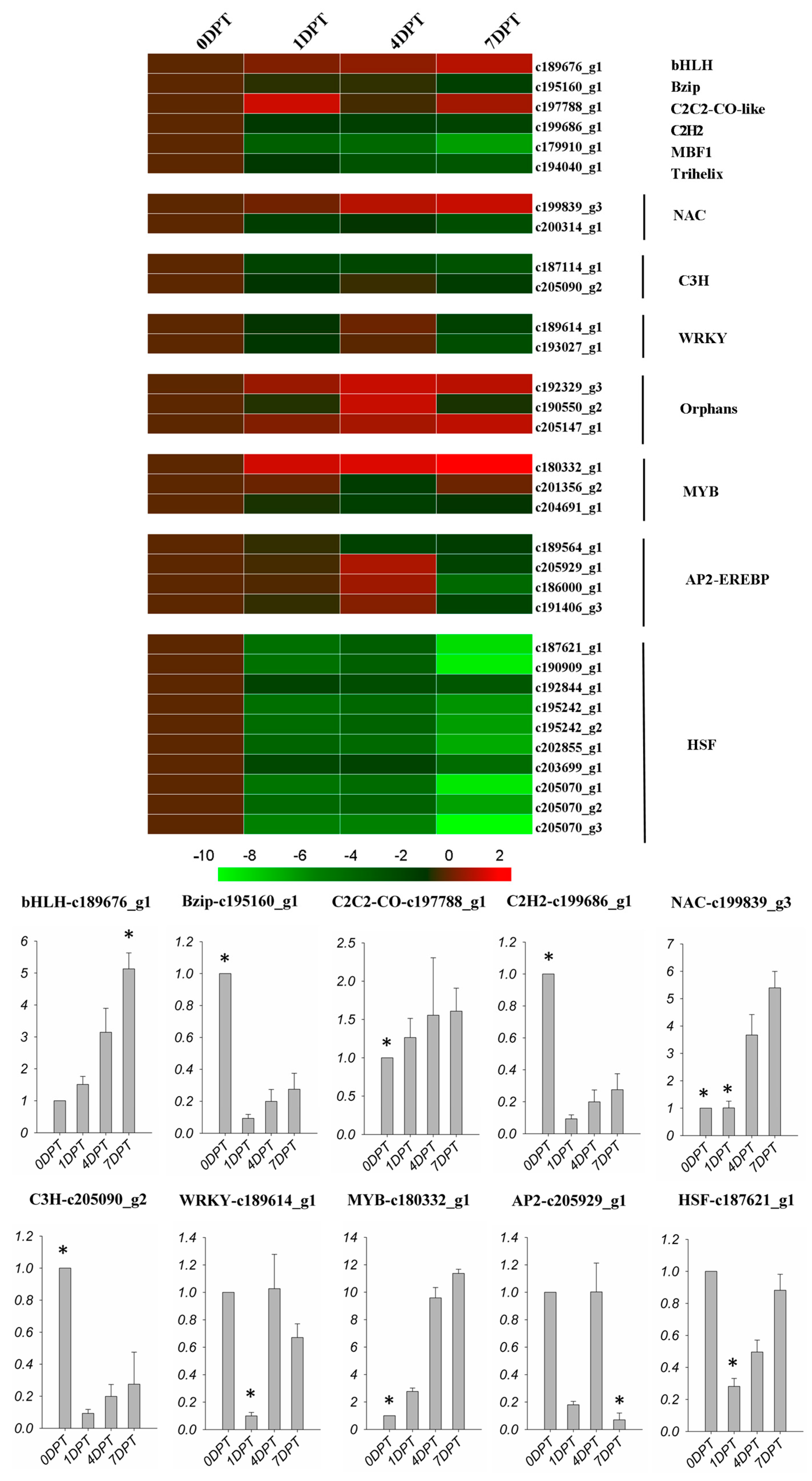
2.6. BD Regulated Transcription Factors
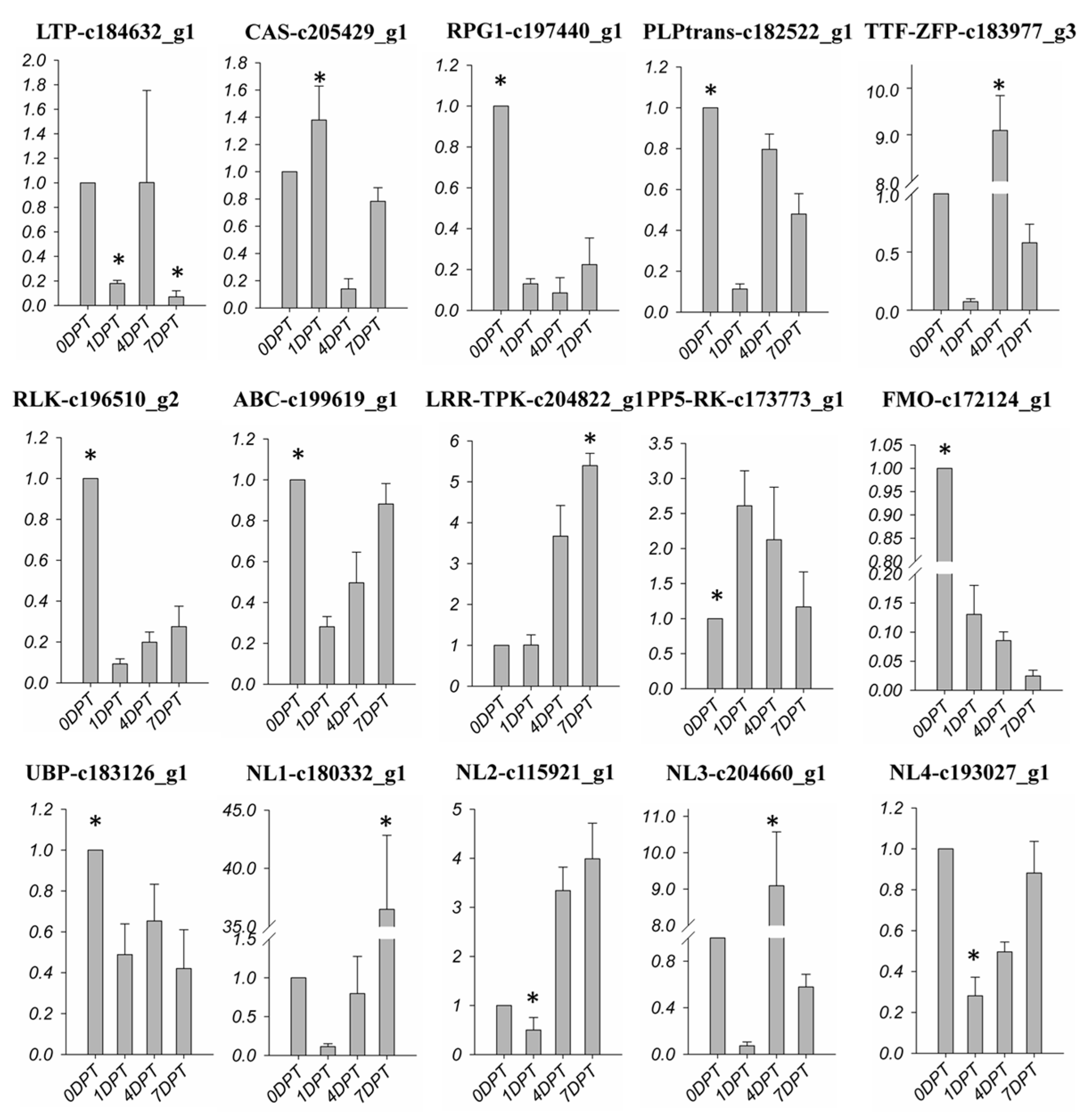
2.7. BD-Regulated R-Gene of Creeping Bentgrass
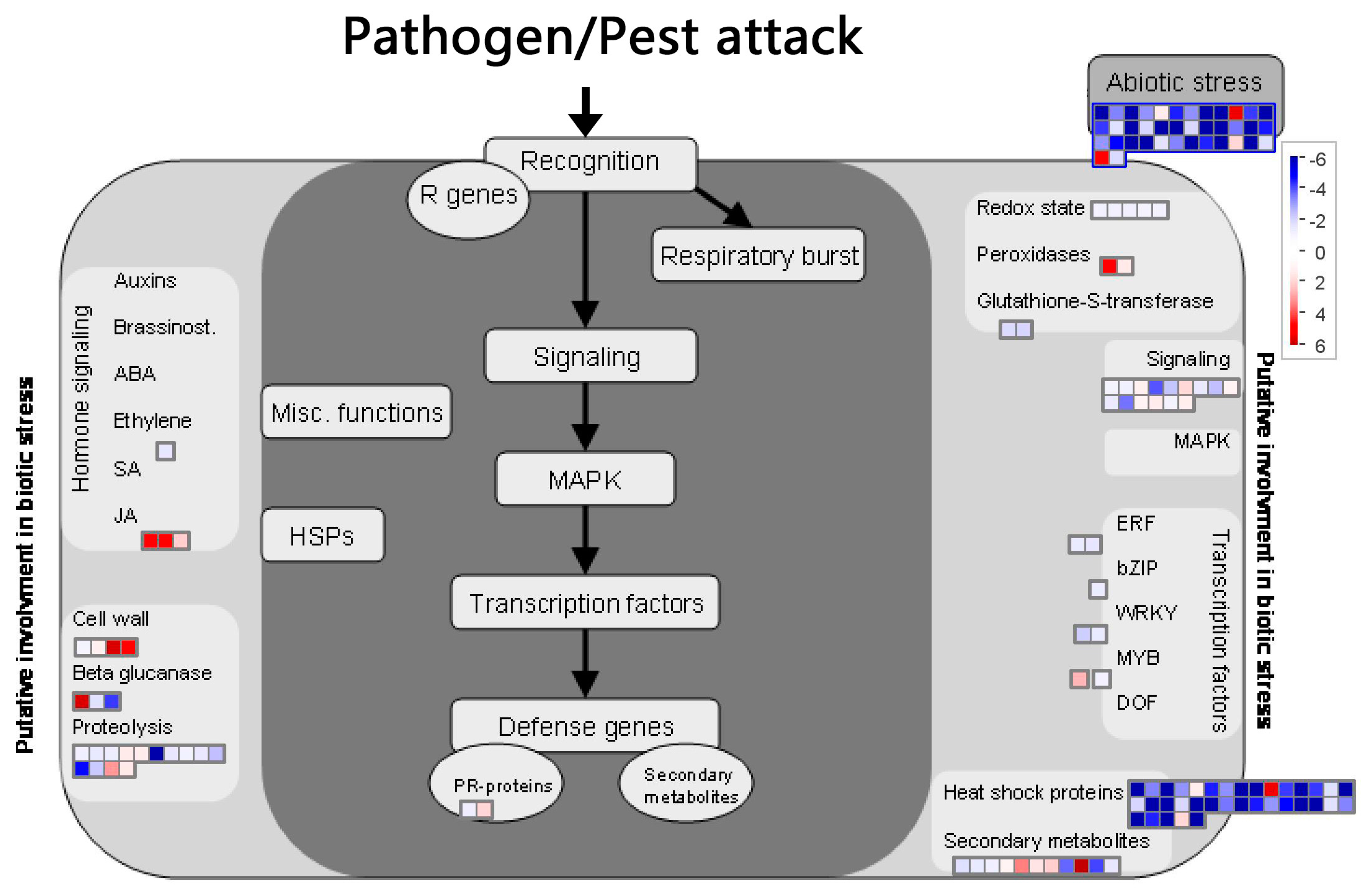
2.8. BD-Induced DEGs Related with Plant Biotic Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Inoculation and Disease Assessments
4.3. Antimicrobial Activity Test
4.4. Sample Preparation and RNA Extraction
4.5. Library Preparation and Transcriptome Sequencing
4.6. Transcriptome Assembly and Annotation
4.7. Transcript Expression Analysis
4.8. Statistical Analysis of Data
5. Conclusions
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of Interest
References
- Kloepper, J.W.; Schroth, M.N.; Miller, T.D. Effects of rhizosphere colonization by plant growth-promoting rhizobacteria on potato plant development and yield. Phytopathology 1980, 70, 1078–1082. [Google Scholar] [CrossRef]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W. Plant growth-promoting rhizobacteria as biological control agents. In Soil Microbial Ecology: Applications in Agricultural and Environmental Management; Metting, F.B., Jr., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1992; pp. 255–274. [Google Scholar]
- Van Loon, L.C.; Bakker, P.A.H.M.; Pierterse, C.M.J. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W.; Rodriguez-Kabana, R.; Zehnder, G.W.; Murphy, J.; Sikora, E.; Fernandez, C. Plant root-bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Aust. J. Plant Pathol. 1999, 28, 27–33. [Google Scholar] [CrossRef]
- Cleyet-Marcel, J.C.; Larcher, M.; Bertrand, H.; Rapior, S.; Pinochet, X. Plant growth enhancement by rhizobacteria. In Nitrogen Assimilation by Plants, Physiological, Biochemical and Molecular Aspects; Morot-Gaudry, J.-F., Ed.; Science Publishers, Inc.: Enfeld, NH, USA, 2001; pp. 185–197. [Google Scholar]
- Alström, S. Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere pseudomonads. J. Gen. Appl. Microbiol. 1991, 37, 495–501. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Bakker, P.A.H.M.; Pieterse, C.M.J. Induction and expression of PGPR-mediated induced resistance against pathogens. IOBC/wprs Bull. 1998, 21, 103–110. [Google Scholar]
- Chen, C.; Belanger, R.R.; Benhamou, N.; Paulitz, T.C. Defense enzymes induced in cucumber roots by treatment with plant growth-promoting rhizobacteria (PGPR) and Pythium aphanidermatum. Phys. Mol. Plant Pathol. 2000, 56, 13–23. [Google Scholar] [CrossRef]
- Leeman, M.; Van Pelt, J.A.; Den Ouden, F.M.; Heinsbroek, M.; Bakker, P.A.H.M.; Schippers, B. Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 1995, 85, 1021–1027. [Google Scholar] [CrossRef]
- Zhang, S.; Reddy, M.S.; Kloepper, J.W. Tobacco growth enhancement and blue mold disease protection by rhizobacteria: Relationship between plant growth promotion and systemic disease protection by PGPR strain 90–166. Plant Soil 2004, 262, 277–288. [Google Scholar] [CrossRef]
- Raupach, G.S.; Liu, L.; Murphy, J.F.; Tuzun, S.; Kloepper, J.W. Induced systemic resistance in cucumber and tomato against cucumber mosaic cucumovirus using plant growth-promoting rhizobacteria (PGPR). Plant Dis. 1996, 80, 891–894. [Google Scholar] [CrossRef]
- Pieterse, C.M.L.; Van Wees, S.C.; Hoffland, E.; Van Pelt, J.A.; Van Loon, L.C. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 1996, 8, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Paré, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Meckes, N.; Pervaiz, Z.H.; Traw, M.B. Microbial interactions in the phyllosphere increase plant performance under herbivore biotic stress. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Ji, H.; Amirullah, A.; Traw, M.B. Pseudomonas syringae pv. tomato DC3000 growth in multiple gene knockouts predicts interactions among hormonal, biotic and abiotic stress responses. Eur. J. Plant Pathol. 2017, 1–8. [Google Scholar] [CrossRef]
- Van der Ent, S.; Van Wees, S.C.; Pieterse, C.M. Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 2009, 70, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Robin, G.P. Systemic signaling during plant defense. Curr. Opin. Plant Biol. 2013, 16, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Kloepper, J.W.; Paré, P.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.S.; Ryu, C.M.; Lee, S.; Park, H.B.; Han, K.S.; Lee, J.H.; Bae, D.W. Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles. Planta 2010, 232, 1355–1370. [Google Scholar] [CrossRef] [PubMed]
- Muyanga, S.C.; Nembaware, V.; Gehring, C. The protection of Pseudomonas aeruginosa wheat seeds treated against powdery mildew and leaf blight correlates with up-regulated expression of a subtilisin-like gene in leaves: Research letter. S. Afr. J. Sci. 2005, 101, 201–204. [Google Scholar]
- Djonovic, S.; Vargas, W.A.; Kolomiets, M.V.; Horndeski, M.; Wiest, A.; Kenerley, C.M. A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 2007, 145, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Lee, S.J.; Moon, J.H.; Park, K.H.; Yang, K.Y.; Cho, B.H.; Kim, K.Y.; Lee, M.C.; Anderson, A.J.; Kim, Y.C. GacS-dependent production of 2R,3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol. Plant Microbe Interact. 2006, 19, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Barco, A.M.; Hsiang, T.; Goodwin, P.H. Induced systemic resistance against three foliar diseases of Agrostis stolonifera by (2R, 3R)-butanediol or an isoparaffin mixture. Annu. Appl. Biol. 2010, 157, 179–189. [Google Scholar] [CrossRef]
- Hussain, S.; Siddique, T.; Saleem, M.; Arshad, M.; Khalid, A. Impact of pesticides on soil microbial diversity, enzymes, and biochemical reactions. Adv. Agron. 2009, 102, 159–200. [Google Scholar]
- Smiley, R.W.; Dernoeden, P.H.; Clarke, B.B. Compendium of Turfgrass Diseases, 3rd ed.; American Phytopathological Society, APS Press: St. Paul, MN, USA, 2005; pp. 11–72. [Google Scholar]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35 (Suppl. 2), W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Vukosavljev, M.; Esselink, G.D.; van’t Westende, W.P.C.; Cox, P.; Visser, R.G.F.; Arens, P.; Smulders, M.J.M. Efficient development of highly polymorphic microsatellite markers based on polymorphic repeats in transcriptome sequences of multiple individuals. Mol. Ecol. Res. 2015, 15, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Annu. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.T.; Zhao, X.; Shang, Q.H.; Wang, Y.; Guo, Z.H.; Zhang, Y.B.; Xie, Z.K.; Wang, R.Y. Comparative Digital Gene Expression Analysis of the Arabidopsis Response to Volatiles Emitted by Bacillus amyloliquefaciens. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Cartieaux, F.; Contesto, C.; Gallou, A.; Desbrosses, G.; Kopka, J.; Taconnat, L.; Renou, J.P.; Touraine, B. Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol. Plant-Microbe Interact. 2008, 21, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Howell, C.R.; Hanson, L.E.; Stipanovic, R.D.; Puckhaber, L.S. Induction of terpenoid synthesis in cotton roots and control of Rhizoctonia solani by seed treatment with Trichoderma virens. Phytopathology 2000, 90, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Shadle, G.L.; Wesley, S.V.; Korth, K.L.; Chen, F.; Lamb, C.; Dixon, R.A. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of l-phenylalanine ammonia-lyase. Phytochemistry 2003, 64, 153–161. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, Y.; Xie, X.; Kim, M.S.; Dowd, S.E.; Paré, P.W. A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 2009, 58, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef] [PubMed]
- Mauch, F.; Mauch-Mani, B.; Boller, T. Antifungal hydrolases in pea tissue II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988, 88, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Stephen, S.; Kazan, K.; Jin, G.; Fan, L.; Taylor, J.; Wang, M.B. Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 2013, 512, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Morán-Diez, E.; Rubio, B.; Domínguez, S.; Hermosa, R.; Monte, E.; Nicolás, C. Transcriptomic response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 2012, 169, 614–620. [Google Scholar] [PubMed]
- Kurth, F.; Feldhahn, L.; Bönn, M.; Herrmann, S.; Buscot, F.; Tarkka, M.T. Large scale transcriptome analysis reveals interplay between development of forest trees and a beneficial mycorrhiza helper bacterium. BMC Genom. 2015, 16, 658. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.S.; Eulgem, T.; Budworth, P.R. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Delessert, C.; Kazan, K.; Wilson, I.W.; Straeten, D.V.D.; Manners, J.; Dennis, E.S.; Dolferus, R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005, 43, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Logemann, E.; Parniske, M.; Hahlbrock, K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. USA 1995, 92, 5905–5909. [Google Scholar] [CrossRef] [PubMed]
- Wu, C. Heat shock transcription factors: Structure and regulation. Annu. Rev. Cell Dev. Biol. 1995, 11, 441–469. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Nomura, H.; Komori, T.; Kobori, M.; Nakahira, Y.; Shiina, T. Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 2008, 53, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Nomura, H.; Komori, T.; Uemura, S.; Kanda, Y.; Shimotani, K.; Nakai, K.; Suwastika, I.N. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012, 3, 926. [Google Scholar] [CrossRef] [PubMed]
- Brueggeman, R.; Rostoks, N.; Kudrna, D.; Kilian, A.; Han, F.; Chen, J.; Kleinhofs, A. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 2002, 99, 9328–9333. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006, 141, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.I.; Chang, P.F.L.; Liu, D.; Narasimhan, M.L.; Raghothama, K.G.; Hasegawa, P.M.; Bressan, R.A. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 1994, 6, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed]
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between domains of a plant NBS–LRR protein in disease resistance-related cell death. EMBO J. 2002, 21, 4511–4519. [Google Scholar] [CrossRef] [PubMed]
- De Young, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.; Creelman, R.A.; Mullet, J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 8675–8679. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I. Full-length transcriptomeassembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K. Method gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Riano-Pachon, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38 (Suppl. 1), D822–D827. [Google Scholar] [CrossRef] [PubMed]
- Sanseverino, W.; Roma, G.; De Simone, M.; Faino, L.; Melito, S.; Stupka, E.; Ercolano, M.R. PRGdb: A bioinformatics platform for plant resistance gene analysis. Nucleic Acids Research 2010, 38 (Suppl. 1), D814–D821. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: All samples are available from the authors. |

| Items | Number of bp/Reads/Transcripts | |
|---|---|---|
| Sequencing and assembly | Total raw reads | 258,606,402 |
| Total clean reads | 252,117,292 | |
| Total transcripts | 466,761 | |
| Total length of transcripts (bp) | 324,797,015 | |
| Transcripts with N50 length (bp) | 1100 | |
| Transcript mean length (bp) | 696 | |
| Total unigenes | 334,212 | |
| Total length of unigenes (bp) | 191,363,312 | |
| Unigenes with N50 length (bp) | 791 | |
| Unigenes mean length (bp) | 573 | |
| Function annotation | NR database | 130,367 |
| NT database | 72,777 | |
| KO database | 41,963 |
| Cluster | Biological Processes | Molecular Functions | Cellular Component |
|---|---|---|---|
| I | Translation | Catalytic activity | Intracellular |
| Phosphorelay signal transduction system | RNA binding | Transcription factor | |
| Metabolic process | Enzyme regulator activity | Membrane | |
| DNA replication | Respiratory chain complex | ||
| Oxidation-reduction process | |||
| II | Lipopolysaccharide core region biosynthetic | Metal ion binding | Transcription factor complex |
| Response to oxidative stress | Hydrolase activity | Component of membrane | |
| Phospholipid catabolic process | Fructose-bisphosphate aldolase activity | ||
| Diacylglycerol kinase activity | |||
| III | Thiamine biosynthetic process | Transcription factor activity | Small-subunit processome |
| Proteolysis | Ubiquitin binding | Peroxidase activity | |
| RNA modification | |||
| IV | Unsaturated fatty acid biosynthetic process | Terpene synthase activity | Fatty acid synthase complex |
| Ferrous iron transport | Serine-type endopeptidase inhibitor activity | Chloroplast | |
| Response to biotic stimulus | Malate synthase activity | Cell wall | |
| Cellular aromatic compound metabolic | Xyloglucosyl transferase activity | ||
| Peroxidase activity | |||
| V | Pathogenesis | Calcium channel activity | Photosystem II oxygen evolving complex |
| Chlorophyll biosynthetic process | Pheromone activity | Magnesium chelatase complex | |
| Protein import into peroxisome matrix | |||
| Nitrogen compound metabolic process | |||
| FtsZ-dependent cytokinesis | |||
| VI | Starch metabolic process | Chitinase activity | Cell junction |
| Sucrose metabolic process | Calmodulin binding | Cell wall | |
| Response to stress | ATPase activator activity | ||
| Glycerolipid metabolic process | |||
| Chitin catabolic process | |||
| VII | Regulation of transcription, DNA-templated | Adenosylcobinamide kinase activity | U1 snRNP |
| Pantothenate biosynthetic process | Aldehyde-lyase activity | Holliday junction helicase complex | |
| Autophagy | |||
| Cobalamin biosynthetic process | |||
| Apoptotic process | |||
| VIII | Cell adhesion | Enzyme inhibitor activity | Plasma membrane |
| Photorespiration | Flavin adenine dinucleotide binding | ||
| Oligosaccharide biosynthetic process | Nutrient reservoir activity | ||
| Galactose metabolic process | |||
| Benzoate metabolic process | |||
| Pectin catabolic process |
| Cluster | KEGG Pathway | q-Value |
|---|---|---|
| I | Ribosome | 4.59 × 10−36 |
| Plant hormone signal transduction | 0.0251 | |
| Glyoxylate and dicarboxylate metabolism | 0.0244 | |
| II | Porphyrin and chlorophyll metabolism | 0.0100 |
| Nitrogen metabolism | 0.0100 | |
| Fructose and mannose metabolism | 0.0100 | |
| Phenylalanine metabolism | 0.0100 | |
| III | Thiamine metabolism | 0.0112 |
| Circadian rhythm-plant | 0.0173 | |
| Amino sugar and nucleotide sugar metabolism | 0.0251 | |
| Carbon metabolism | 0.0453 | |
| IV | Ribosome biogenesis in eukaryotes | 0.0181 |
| Monoterpenoid biosynthesis | 0.0183 | |
| alpha-Linolenic acid metabolism | 0.0181 | |
| Pyruvate metabolism | 0.0181 | |
| Sesquiterpenoid and triterpenoid biosynthesis | 0.0301 | |
| Carotenoid biosynthesis | 0.0357 | |
| V | Protein processing in endoplasmic reticulum | 1.23 × 10−17 |
| Spliceosome | 0.0125 | |
| Cutin, suberine and wax biosynthesis | 0.0485 | |
| Metabolism of xenobiotics by cytochrome P450 | 0.0157 | |
| MAPK signaling pathway | 0.0385 | |
| Arachidonic acid metabolism | 0.0459 | |
| Phenylalanine metabolism | 0.0732 | |
| VI | NOD-like receptor signaling pathway | 8.38 × 10−6 |
| Glutathione metabolism | 0.0304 | |
| Ubiquinone and other terpenoid-quinone biosynthesis | 0.0304 | |
| Phenylpropanoid biosynthesis | 0.0340 | |
| Cyanoamino acid metabolism | 0.0484 | |
| VII | Photosynthesis - antenna proteins | 5.97 × 10−7 |
| NOD-like receptor signaling pathway | 6.25 × 10−5 | |
| Carbon fixation in photosynthetic organisms | 0.0061 | |
| VIII | NOD-like receptor signaling pathway | 9.69 × 10−5 |
| Plant-pathogen interaction | 0.0012 | |
| Photosynthesis | 0.0301 | |
| Brassinosteroid biosynthesis | 0.0185 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Niu, K.; Huang, B.; Liu, W.; Ma, H. Transcriptional Responses of Creeping Bentgrass to 2,3-Butanediol, a Bacterial Volatile Compound (BVC) Analogue. Molecules 2017, 22, 1318. https://doi.org/10.3390/molecules22081318
Shi Y, Niu K, Huang B, Liu W, Ma H. Transcriptional Responses of Creeping Bentgrass to 2,3-Butanediol, a Bacterial Volatile Compound (BVC) Analogue. Molecules. 2017; 22(8):1318. https://doi.org/10.3390/molecules22081318
Chicago/Turabian StyleShi, Yi, Kuiju Niu, Bingru Huang, Wenhui Liu, and Huiling Ma. 2017. "Transcriptional Responses of Creeping Bentgrass to 2,3-Butanediol, a Bacterial Volatile Compound (BVC) Analogue" Molecules 22, no. 8: 1318. https://doi.org/10.3390/molecules22081318





