Molecular Characterization and Expression Profiles of Sp-uchl3 and Sp-uchl5 during Gonad Development of Scylla paramamosain
Abstract
:1. Introduction
2. Results
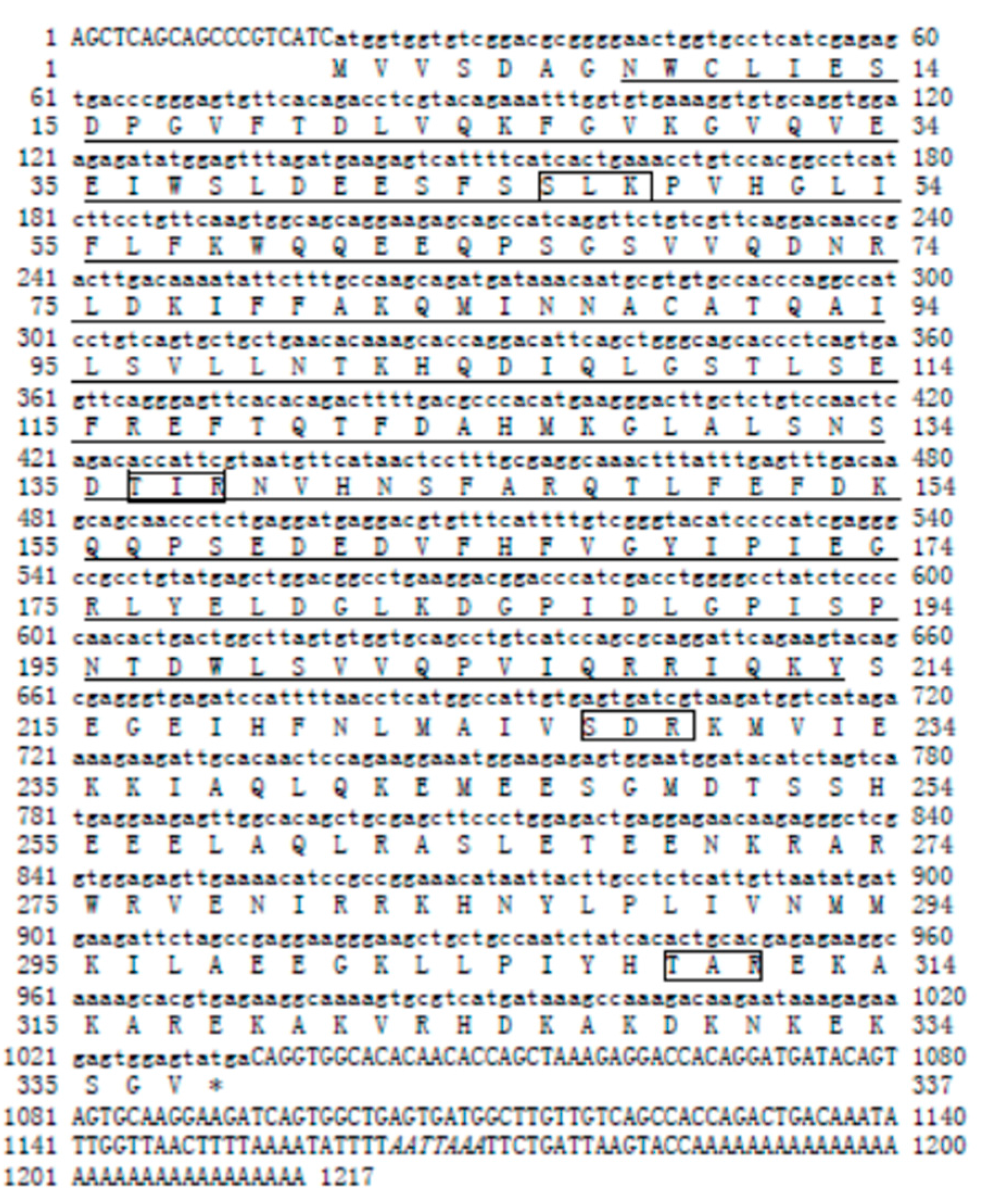
2.1. Molecular Characterization of Sp-uchl3 and Sp-uchl5
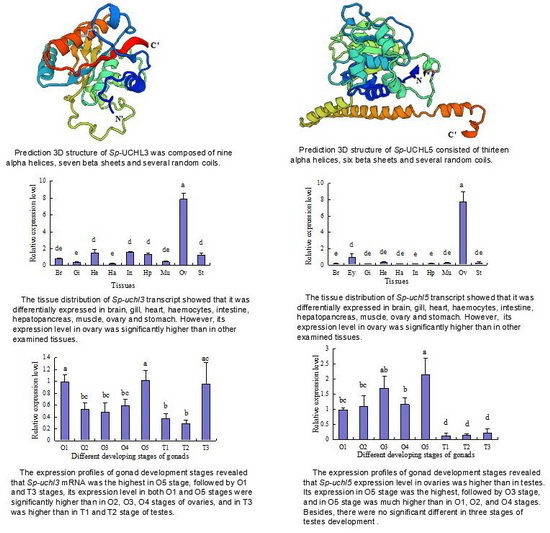
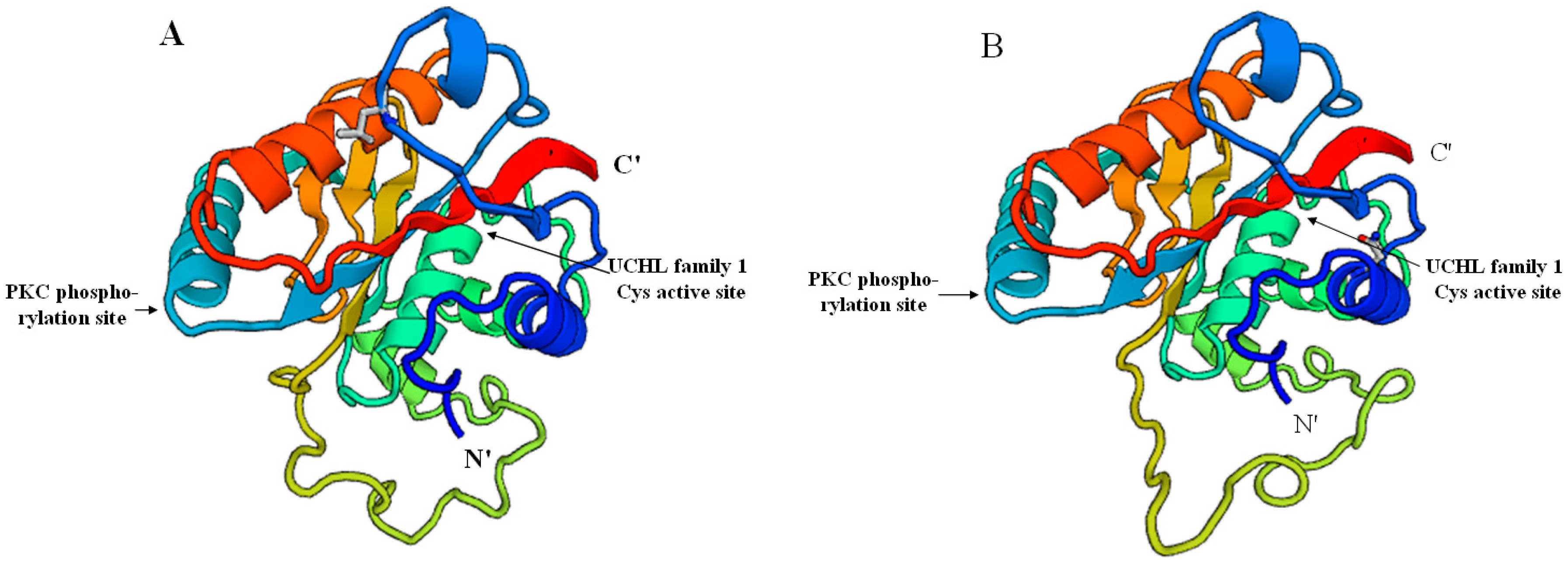
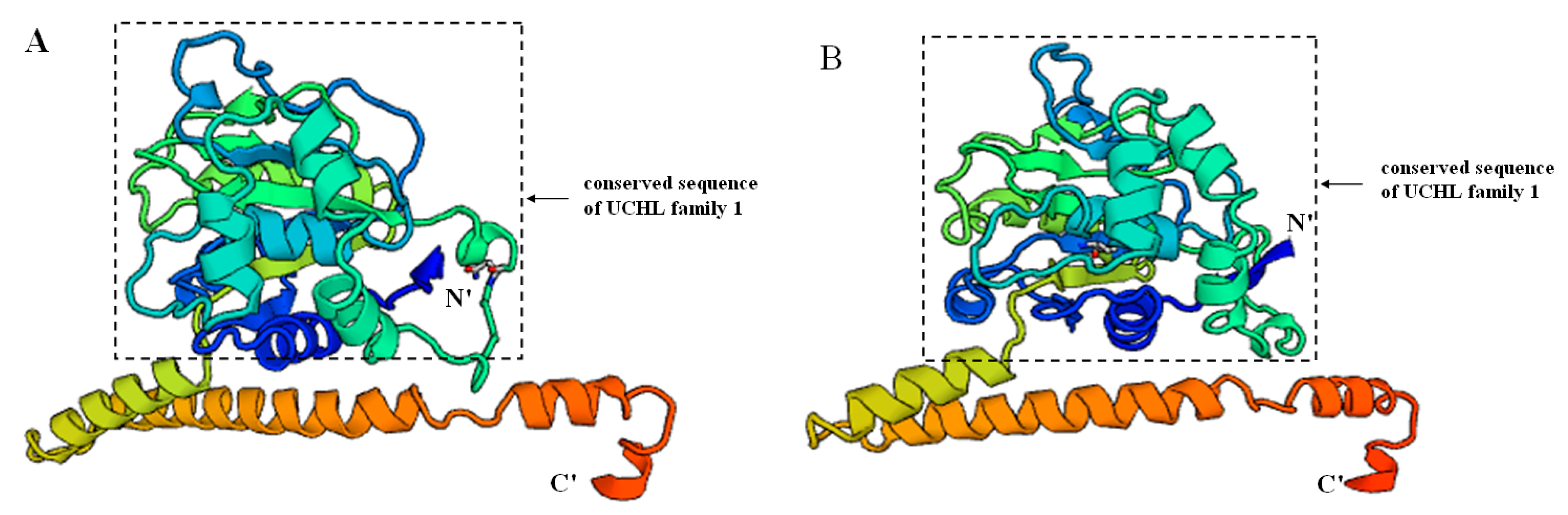
2.2. Prediction of Sp-UCHL3 and Sp-UCHL5 Three-Dimensional (3D) Structure
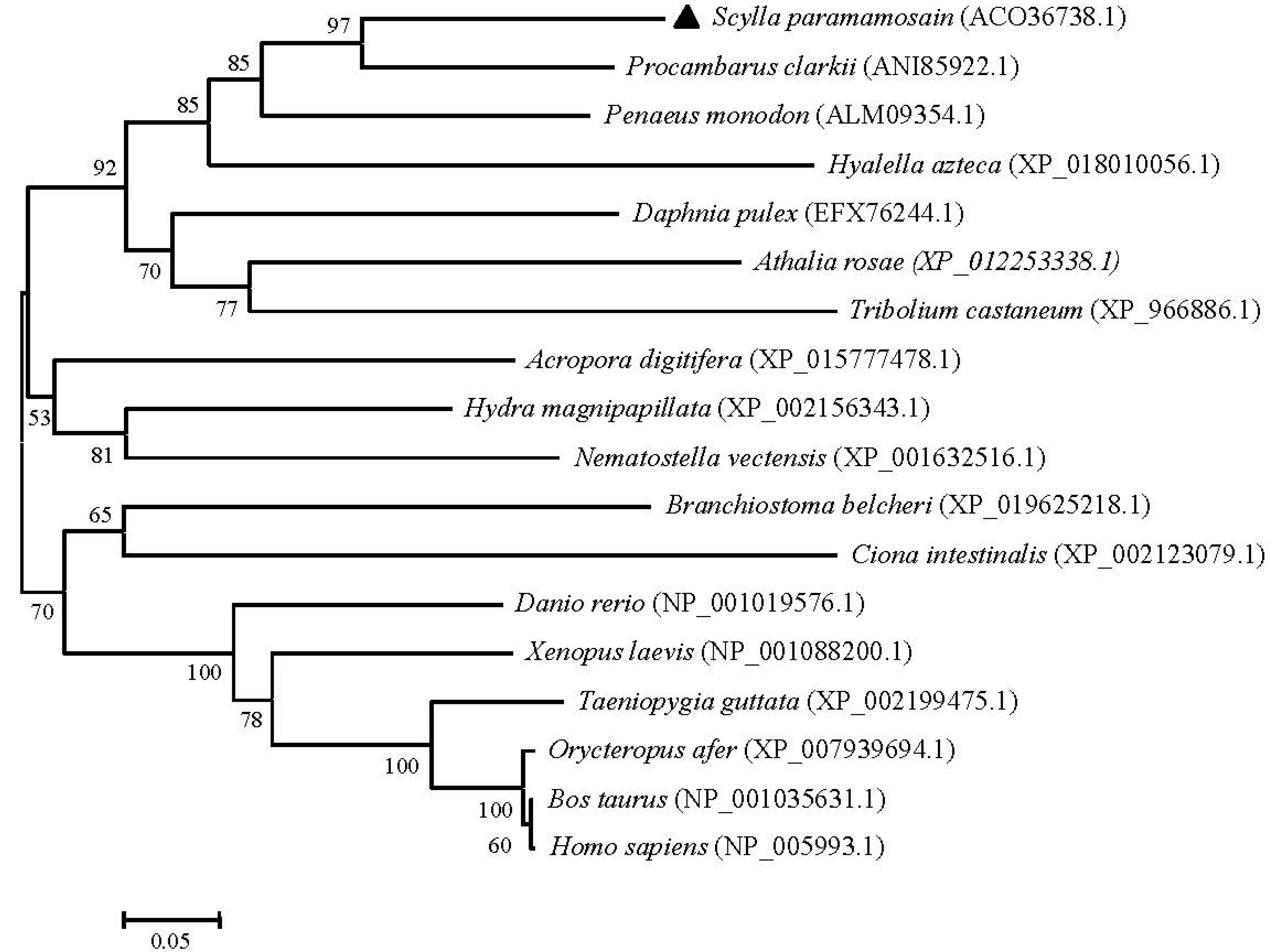
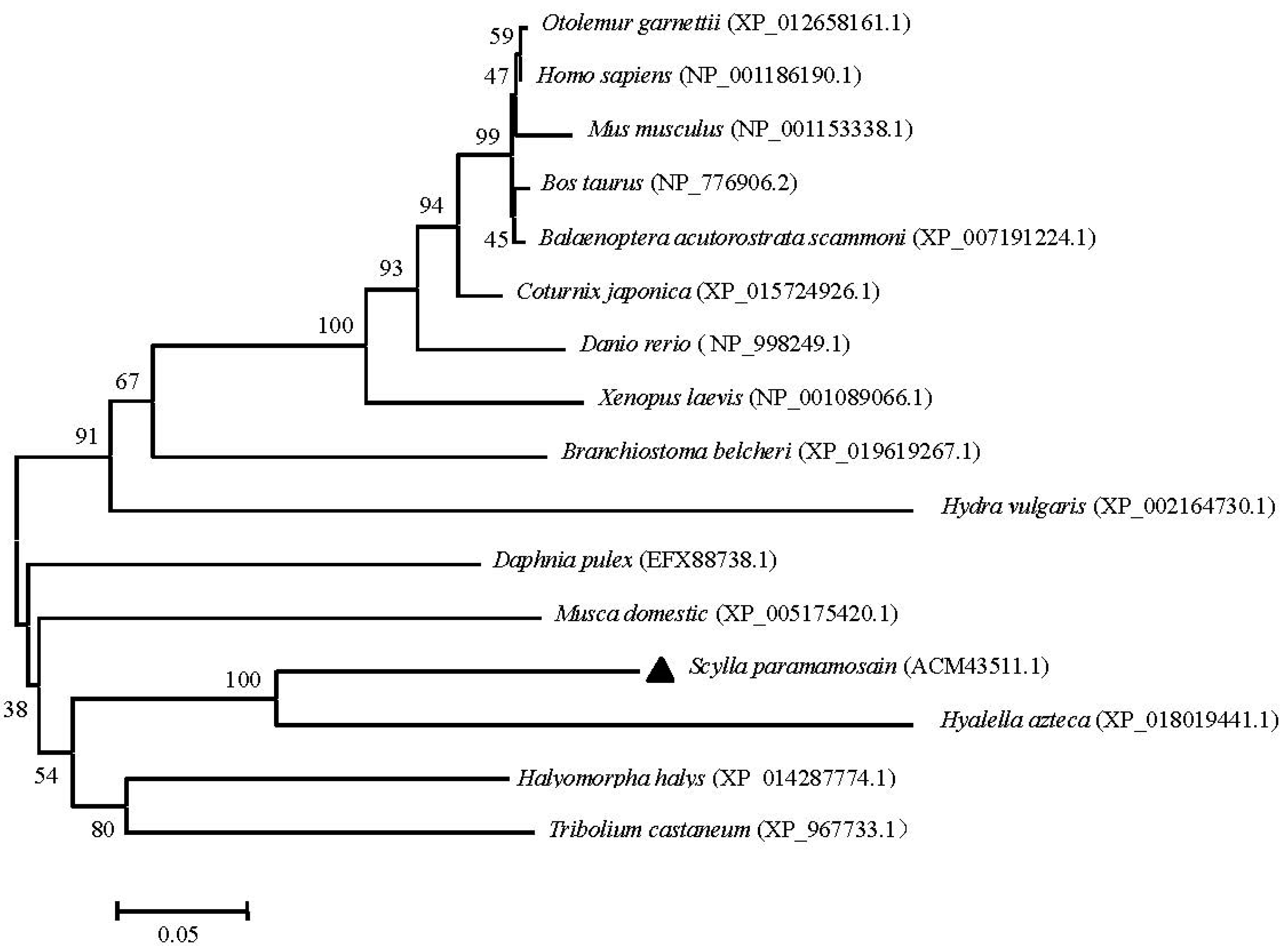
2.3. Phylogenetic and Homology Analyses of Sp-UCHL3 and Sp-UCHL5
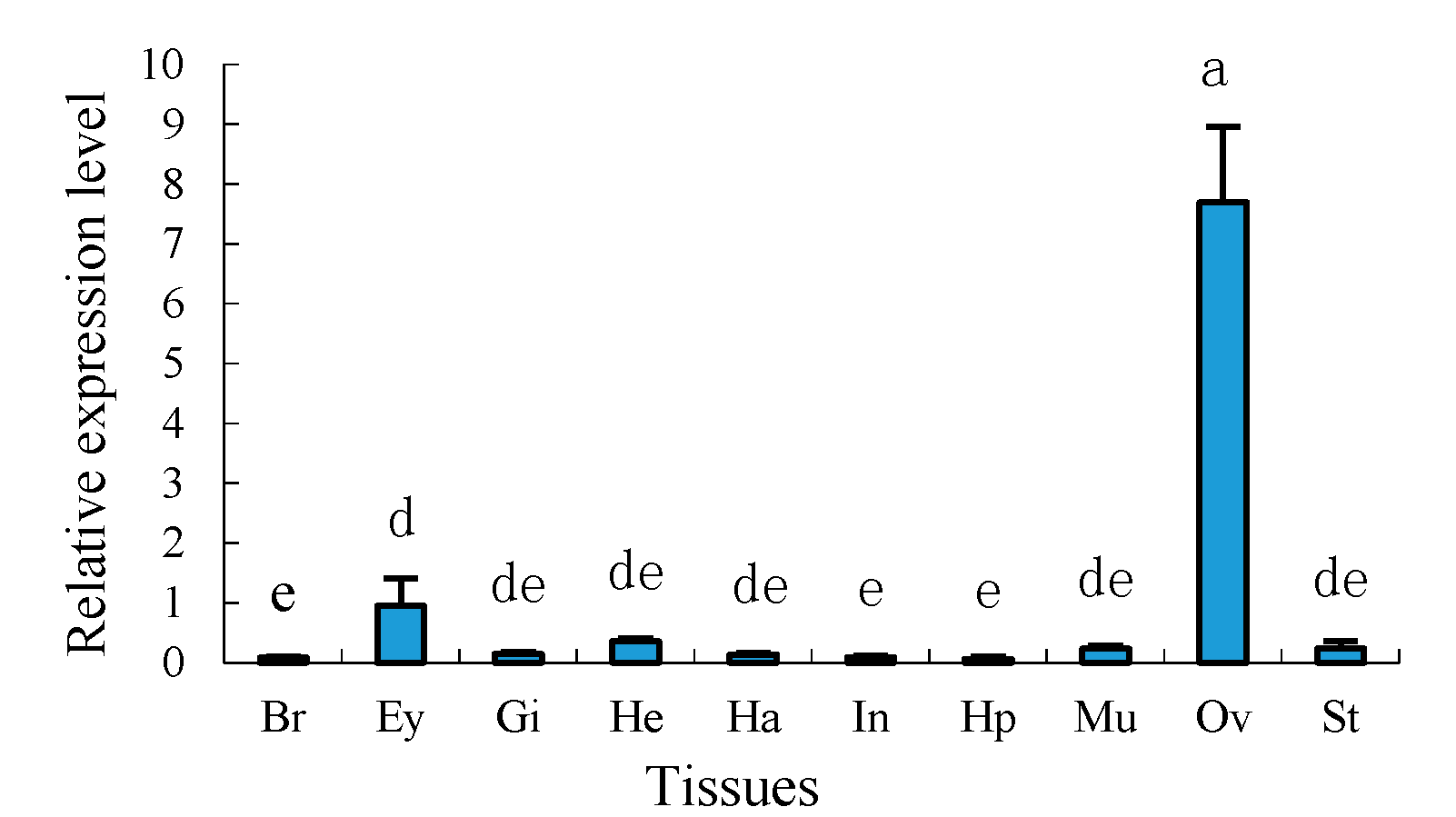
2.4. Tissue Distribution of Sp-uchl3 and Sp-uchl5 Transcripts in Female Crab
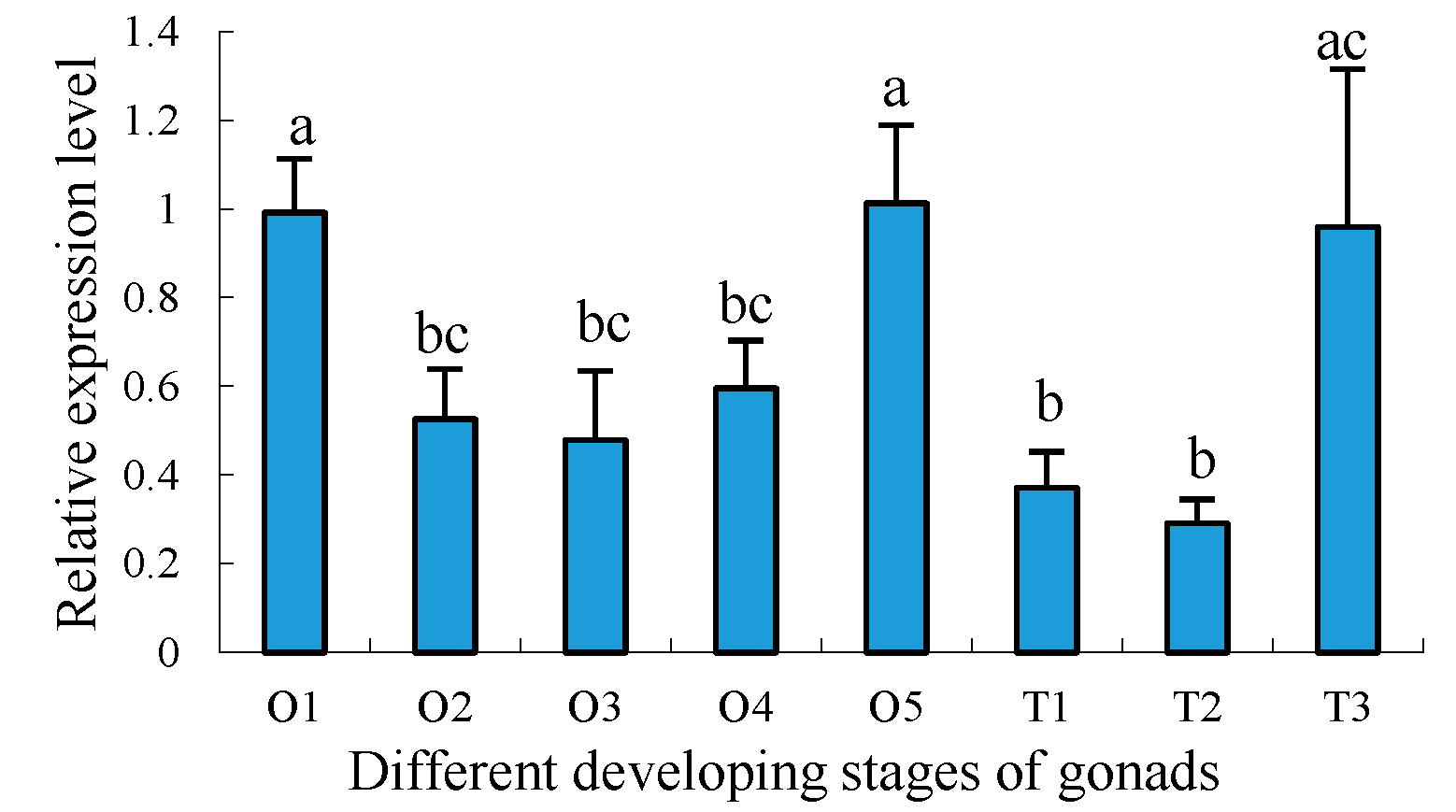
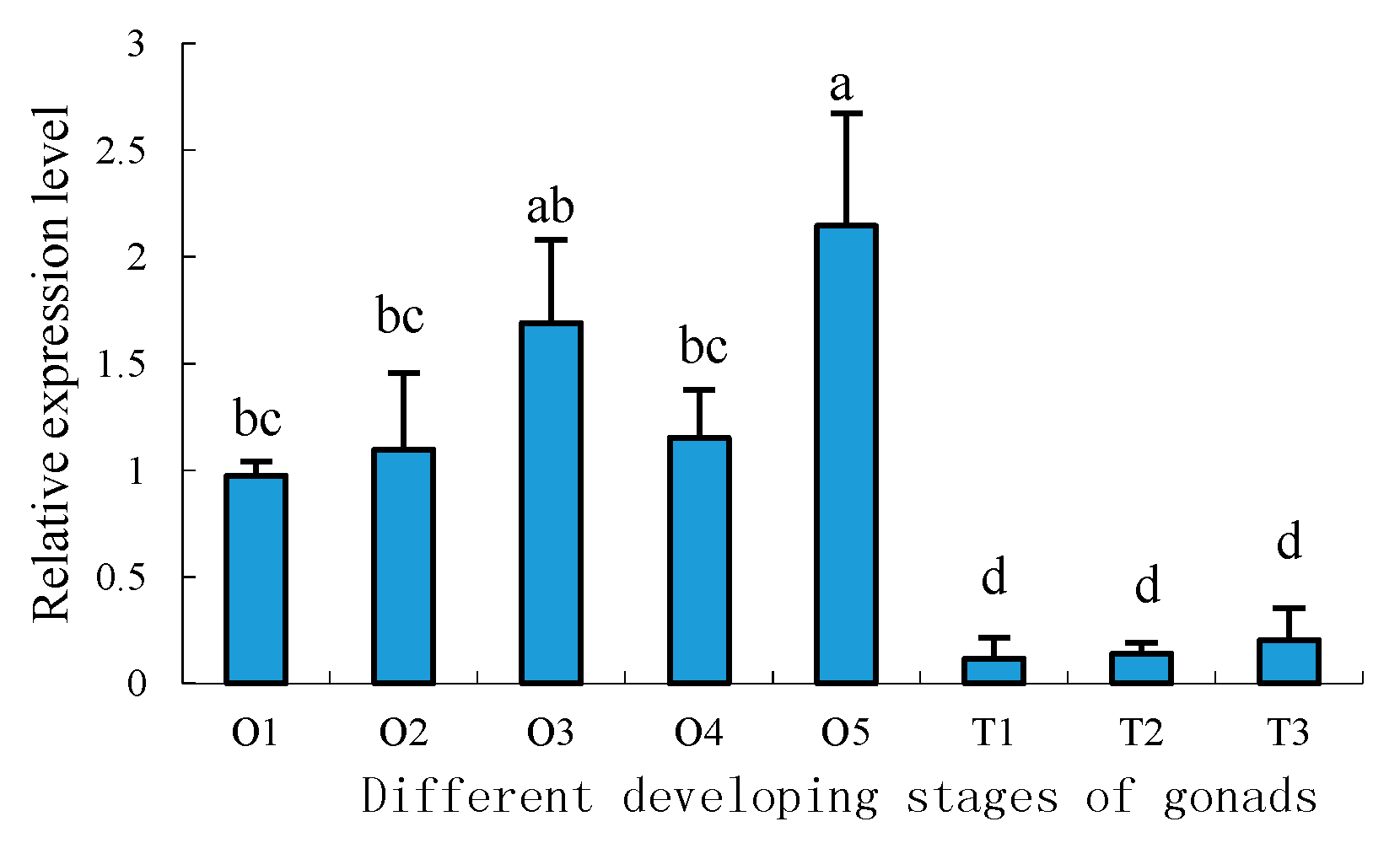
2.5. Expression Profiles of Sp-uchl3 and Sp-uchl5 Transcripts in the Different Stages of Gonad Development
3. Discussion
4. Materials and Methods
4.1. Animals and Tissue Collection
4.2. RNA Isolation and cDNA Synthesis
4.3. cDNA Cloning for Sp-uchl3 and Sp-uchl5
4.4. Bioinformatics Analysis of Sequences
4.5. RT-qPCR
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ye, H.; Tao, Y.; Wang, G.; Lin, Q.; Chen, X.; Li, S. Experimental nursery culture of the mud crab Scylla paramamosain (Estampador) in China. Aquac. Int. 2011, 19, 313–321. [Google Scholar] [CrossRef]
- Li, S.J.; Wang, G.Z. Biology & Artificial Cultivation of Mud Crab Scylla serrata (Forskal 1775); Xiamen University Press: Xiamen, China, 2007; pp. 611–705. (In Chinese) [Google Scholar]
- Wang, G.Z.; Ye, H.H.; Li, S.J. Status and suggestions of mud crab aquaculture in Fujian Province. J. Fujian Fish. 2012, 34, 87–90. (In Chinese) [Google Scholar]
- Zhu, X.; Zou, Q.; Li, S.; Wang, G. Aquaculture of the Mud Crab (Scylla spp.) in Inshore Shrimp Ponds throughout Southern China. J. Xiamen Univ. (Nat. Sci.) 2006, 45, 256–260. (In Chinese) [Google Scholar]
- Gong, J.; Huang, C.; Shu, L.; Bao, C.; Huang, H.; Ye, H.; Zeng, C.; Li, S. The retinoid X receptor from mud crab: New insights into its roles in ovarian development and related signaling pathway. Sci. Rep. 2016, 6, 23654. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Bao, C.; Huang, H.; Ye, H.; Li, S. The mechanism of regulation of ovarian maturation by red pigment concentrating hormone in the mud crab Scylla paramamosain. Anim. Reprod. Sci. 2016, 164, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Yang, Y.; Huang, H.; Ye, H. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: Transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 2015, 5, 17055. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zhang, Z.; Wang, Y.; Han, K.; Fu, M.; Lin, P.; Jia, X. EST analysis on the gonad development related organs and microarray screen for differentially expressed genes in mature ovary and testis of Scylla paramamosain. Comp. Biochem. physiol. Part D Genom. Proteom. 2011, 6, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, X.; Zou, Z.; Jia, X.; Wang, Y.; Zhang, Z. Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab (Scylla paramamosain). BMC Genom. 2014, 15, 585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Han, K.; Zou, Z.; Zhang, Z. A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Mol. Biol. Rep. 2012, 39, 4327–4335. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Chen, Y.; Zou, Z.; Lin, P.; Wang, Y.; Zhang, Z. Characterization and expression profile of Vitellogenin gene from Scylla paramamosain. Gene 2013, 520, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Wang, Y.; Zou, Z.; Fu, M.; Lin, P.; Zhang, Z. Erk2 in ovarian development of green mud crab Scylla paramamosain. DNA Cell Biol. 2012, 31, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Dai, Y.; Zou, Z.; Fu, M.; Wang, Y.; Zhang, Z. Molecular characterization and expression profiles of cdc2 and cyclin B during oogenesis and spermatogenesis in green mud crab (Scylla paramamosain). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 163, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Han, K.; Yan, S.; Zou, Z.; Zhang, Z.; Wang, Y. Molecular characterization and expression profiles of Sp-Ub during gonad development in Scylla paramamosain. J. Fish. Sci. China 2012, 19, 946–955. (In Chinese) [Google Scholar] [CrossRef]
- Dai, Y.; Han, K.; Zou, Z.; Yan, S.; Wang, Y.; Zhang, Z. SUMO-1 of mud crab (Scylla paramamosain) in gametogenesis. Gene 2012, 503, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, D.; He, F.C. Bioinformatics advances in protein ubiquitination. Yi Chuan 2013, 35, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003, 19, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Wickliffe, K.; Williamson, A.; Jin, L.; Rape, M. The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 2009, 109, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Chen, C.; Pan, J.; Xu, J.; Zhou, Z.G.; Wang, C.Y. The Ubiquitin Proteasome Pathway (UPP) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. Int. J. Clin. Exp. Pathol. 2012, 5, 726–738. [Google Scholar] [PubMed]
- Mukhopadhyay, D.; Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Grice, G.L.; Nathan, J.A. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016, 73, 3497–3506. [Google Scholar] [CrossRef] [PubMed]
- Baarends, W.M.; Roest, H.P.; Grootegoed, J.A. The ubiquitin system in gametogenesis. Mol. Cell. Endocrinol. 1999, 151, 5–16. [Google Scholar] [CrossRef]
- Bose, R.; Manku, G.; Culty, M.; Wing, S.S. Ubiquitin-proteasome system in spermatogenesis. Adv. Exp. Med. Biol. 2014, 759, 181–213. [Google Scholar] [PubMed]
- Wang, H.M.; Zhu, C. Ubiquitin proteasome Pathway in Reproductive Tissues. Prog. Biochem. Biophys. 2002, 29, 31–34. (In Chinese) [Google Scholar]
- He, W.; Liu, A.; Huang, H. The research process for ubiquitin proteasome pathway in reproductive. J. Int. Reprod. Health/Fam. Plan. 2008, 27, 138–141. (In Chinese) [Google Scholar]
- Bebington, C.; Doherty, F.J.; Fleming, S.D. The possible biological and reproductive functions of ubiquitin. Hum. Reprod. Update 2001, 7, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Rose, I.A. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J. Biol. Chem. 1985, 260, 7903–7910. [Google Scholar] [PubMed]
- Ciechanover, A. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 1998, 17, 7151–7160. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.H.; Liu, H. Life and death in the trash heap: The ubiquitin proteasome pathway and UCHL1 in brain aging, neurodegenerative disease and cerebral Ischemia. Ageing Res. Rev. 2017, 34, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J. The new function of two ubiquitin C-terminal hydrolase isozymes as reciprocal modulators of germ cell apotosis. Exp. Anim. 2007, 56, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Kon, Y.; Endoh, D.; Iwanaga, T. Expression of protein gene product 9.5, a neuronal ubiquitin C-terminal hydrolase, and its developing change insertoli cells of mouse testis. Mol. Reprod. Dev. 1999, 54, 333–341. [Google Scholar] [CrossRef]
- Sekiguchi, S.; Kwon, J.; Yoshida, E.; Hamasaki, H.; Ichinose, S.; Hideshima, M.; Kuraoka, M.; Takahashi, A.; Ishii, Y.; Kyuwa, S.; et al. Localization of ubiquitin C-terminal hydrolase L1 in mouse ova and its function in the plasma membrane to block polyspermy. Am. J. Pathol. 2006, 169, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.G.; Kong, W.H.; Yan, S.; Gu, Z.; Zuo, J.K. The functions in the progesterone-induced oocyte maturation of toad ubiquitin carboxyl-terminal hydrolase (tUCH) is independent of its UCH activity. Shi Yan Sheng Wu Xue Bao 2003, 36, 105–112. [Google Scholar] [PubMed]
- Mochida, K.; Matsubara, T.; Kudo, H.; Andoh, T.; Ueda, H.; Adachi, S.; Yamauchi, K. Molecular cloning and immunohistochemical localization of ubiquitin C-Terminal hydrolase expressed in testis of a teleost, the Nile Tilapia, Oreochromis niloticus. J. Exp. Zool. 2002, 293, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhang, Z.; Wang, Y.; Zou, Z. Cyclin-CDK-CKI and UPP participate in the regulation of reproduction and the progression of gonad development in crustacean. Biotechnol. Bull. 2010, 7, 48–54. (In Chinese) [Google Scholar]
- Kim, J.H.; Park, K.C.; Chung, S.S.; Bang, O.; Chung, C.H. Deubiquitinating enzymes as cellular regulators. J. Biochem. 2003, 134, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Nakasone, M.A.; Lewis, T.A.; Walker, O.; Thakur, A.; Mansour, W.; Castañeda, C.A.; Goeckeler-Fried, J.L.; Parlati, F.; Chou, T.F.; Hayat, O.; et al. Structural Basis for the Inhibitory Effects of Ubistatins in the Ubiquitin-Proteasome Pathway. Structure 2017, 25, 1839–1855. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Suresh, B.; Baek, K.H. The role of deubiquitinating enzymes in apoptosis. Cell. Mol. Life Sci. 2011, 68, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.B.; Morgan, D.O. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J. Biol. Chem. 2011, 286, 45186–45196. [Google Scholar] [CrossRef] [PubMed]
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 2014, 1843, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Larsen, C.N.; Krantz, B.A.; Wilkinson, K.D. Substrate specificity of deubiquitinating enzymes: Ubiquitin C-terminal hydrolases. Biochemistry 1998, 37, 3358–3368. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fu, D.; Shen, X.Z. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta 2010, 1806, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.C.; Riddle, S.M.; Cohen, R.E.; Hill, C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999, 18, 3877–3887. [Google Scholar] [CrossRef] [PubMed]
- Popp, M.W.; Artavanis-Tsakonas, K.; Ploegh, H.L. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J. Biol. Chem. 2009, 284, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, L.J.; Semenova, E.; Levorse, J.M.; Tilghman, S.M. Expression and functional analysis of Uch-L3 during mouse development. Mol. Cell. Biol. 2000, 20, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Boudreaux, D.A.; Maiti, T.K.; Davies, C.W.; Das, C. Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc. Natl. Acad. Sci. USA 2010, 107, 9117–9122. [Google Scholar] [CrossRef] [PubMed]
- Bishop, P.; Rocca, D.; Henley, J.M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): Structure, distribution and roles in brain function and dysfunction. Biochem. J. 2016, 473, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Papaevgeniou, N.; Chondrogianni, N. The ubiquitin proteasome system in Caenorhabditis elegans and its regulation. Redox Biol. 2014, 2, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.R.; Zhang, Y.H.; Liu, S.; Song, A.X.; Hu, H.Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 2012, 441, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Mtango, N.R.; Sutovsky, M.; Vandevoort, C.A.; Latham, K.E.; Sutovsky, P. Essential role of ubiquitin C-terminal hydrolases UCHL1 and UCHL3 in mammalian oocyte maturation. J. Cell. Physiol. 2012, 227, 2022–2029. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.J.; Sutovsky, M.; Song, W.H.; Sutovsky, P. Protein deubiquitination during oocyte maturation influences sperm function during fertilisation, antipolyspermy defense and embryo development. Reprod. Fertil. Dev. 2015, 27, 1154–1167. [Google Scholar] [CrossRef] [PubMed]
- Laney, J.D.; Hochstrasser, M. Substrate targeting in the ubiquitin system. Cell 1999, 97, 427–430. [Google Scholar] [CrossRef]
- Murray, A.; Hunt, T. The Cell Cycle: An Introduction; Oxford University Press: New York, NY, USA, 1993. [Google Scholar]
- Mtango, N.R.; Latham, K.E.; Sutovsky, P. Deubiquitinating enzymes in oocyte maturation, fertilization and preimplantation embryo development. Adv. Exp. Med. Biol. 2014, 759, 89–110. [Google Scholar] [PubMed]
- Ellederova, Z.; Halada, P.; Man, P.; Kubelka, M.; Motlik, J.; Kovarova, H. Protein patterns of pig oocytes during in vitro maturation. Biol. Reprod. 2004, 71, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.G.; Kong, W.H.; Zhang, Y.J.; Yan, S.; Lu, J.N.; Gu, Z.; Lin, F.; Tso, J.K. A novel ubiquitin carboxyl terminal hydrolase is involved in toad oocyte maturation. Cell Res. 2002, 12, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Kodama, K.; Kurokura, H. Ovarian development of the mud crab Scylla paramamosain in a tropical mangrove swamp, Thailand. J. Sci. Res. 2010, 2, 380–389. [Google Scholar] [CrossRef]
- Tsukimura, B. Crustacean vitellogenesis: Its role in oocyte development. Am. Zool. 2001, 41, 465–476. [Google Scholar] [CrossRef]
- Subramoniam, T. Mechanisms and control of vitellogenesis in crustaceans. Fish. Sci. 2011, 77, 1–21. [Google Scholar] [CrossRef]
- Suresh, B.; Lee, J.; Hong, S.H.; Kim, K.S.; Ramakrishna, S. The role of deubiquitinating enzymes in spermatogenesis. Cell. Mol. Life Sci. 2015, 72, 4711–4720. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, T.; Hu, L.; Cheng, Z.; Li, M. Ubiquitin Carboxy-Terminal Hydrolase L3 Correlates with Human Sperm Count, Motility and Fertilization. PLoS ONE 2016, 11, e0165198. [Google Scholar]
- Kwon, J.; Wang, Y.L.; Setsuie, R.; Sekiguchi, S.; Sakurai, M.; Sato, Y.; Lee, W.W.; Ishii, Y.; Kyuwa, S.; Noda, M.; et al. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol. Reprod. 2004, 71, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Kikuchi, T.; Setsuie, R.; Ishii, Y.; Kyuwa, S.; Yoshikawa, Y. Characterization of the testis in congenitally ubiquitin carboxy-terminal hydrolase-1 (Uch-L1) defective (gad) mice. Exp. Anim. 2003, 52, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Mochida, K.; Wang, Y.L.; Sekiguchi, S.; Sankai, T.; Aoki, S.; Ogura, A.; Yoshikawa, Y.; Wada, K. Ubiquitin C-terminal hydrolase L-1 is essential for the early apoptotic wave of germinal cells and for sperm quality control during spermatogenesis. Biol. Reprod. 2005, 73, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, R.S.; Mok, H.O.; Wang, Y.; Poon, W.W.; Cheng, S.H.; Kong, R.Y. Isolation, characterization and expression analysis of a hypoxia-responsive glucose transporter gene from the grass carp, Ctenopharyngodon idellus. Eur. J. Biochem. 2003, 270, 3010–3017. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of Sp-uchl3 and Sp-uchl5 cDNAs are available from the authors. |


| Primer | Primer Sequence (5′-3′) | Purpose |
|---|---|---|
| uchl3-5′ outer | CACAGCAAAGCGAGACTCATCAGG | 5′ RACE |
| uchl3-5′ inner | GGCAGCATCAGCAAGAACAGAGTC | |
| uchl3-3′ outer | GCTTGTGGCACGGTGGCTTTG | 3′ RACE |
| uchl3-3′ inner | GCGAAGCCCAAGTCAACG | |
| RT-uchl3-F | GCGAAGCCCAAGTCAACG | qRT-PCR |
| RT-uchl3-R | CCACCACAGCAAAGCGAGA | |
| uchl5-5′ outer | GCCGTGGACAGGTTTCAGTGATG | 5′ RACE |
| uchl5-5′ inner | CTTCCACCTGCACACCTTTCACA | |
| uchl5-3′ outer | GCAGCCATCAGGTTCTGTCGTTCA | 3′ RACE |
| uchl5-3′ inner | GCCCACATGAAGGGACTTGCTC | |
| RT-uchl5-F | GCCATCAGGTTCTGTCGTTC | RT-qPCR |
| RT-uchl5-R | CATGTGGGCGTCAAAAGTCT | |
| RT-18S-F | ATGATAGGGATTGGGGTTTGC | RT-qPCR |
| RT-18S-R | AAGAGTGCCAGTCCGAAGG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.; Dai, Y.; Zhang, Z.; Zou, Z.; Wang, Y. Molecular Characterization and Expression Profiles of Sp-uchl3 and Sp-uchl5 during Gonad Development of Scylla paramamosain. Molecules 2018, 23, 213. https://doi.org/10.3390/molecules23010213
Han K, Dai Y, Zhang Z, Zou Z, Wang Y. Molecular Characterization and Expression Profiles of Sp-uchl3 and Sp-uchl5 during Gonad Development of Scylla paramamosain. Molecules. 2018; 23(1):213. https://doi.org/10.3390/molecules23010213
Chicago/Turabian StyleHan, Kunhuang, Yanbin Dai, Ziping Zhang, Zhihua Zou, and Yilei Wang. 2018. "Molecular Characterization and Expression Profiles of Sp-uchl3 and Sp-uchl5 during Gonad Development of Scylla paramamosain" Molecules 23, no. 1: 213. https://doi.org/10.3390/molecules23010213
APA StyleHan, K., Dai, Y., Zhang, Z., Zou, Z., & Wang, Y. (2018). Molecular Characterization and Expression Profiles of Sp-uchl3 and Sp-uchl5 during Gonad Development of Scylla paramamosain. Molecules, 23(1), 213. https://doi.org/10.3390/molecules23010213