Pichia cactophila and Kluyveromyces lactis are Highly Efficient Microbial Cell Factories of Natural Amino Acid-Derived Aroma Compounds
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of the Yeast Monocultures and Taxonomic Classification
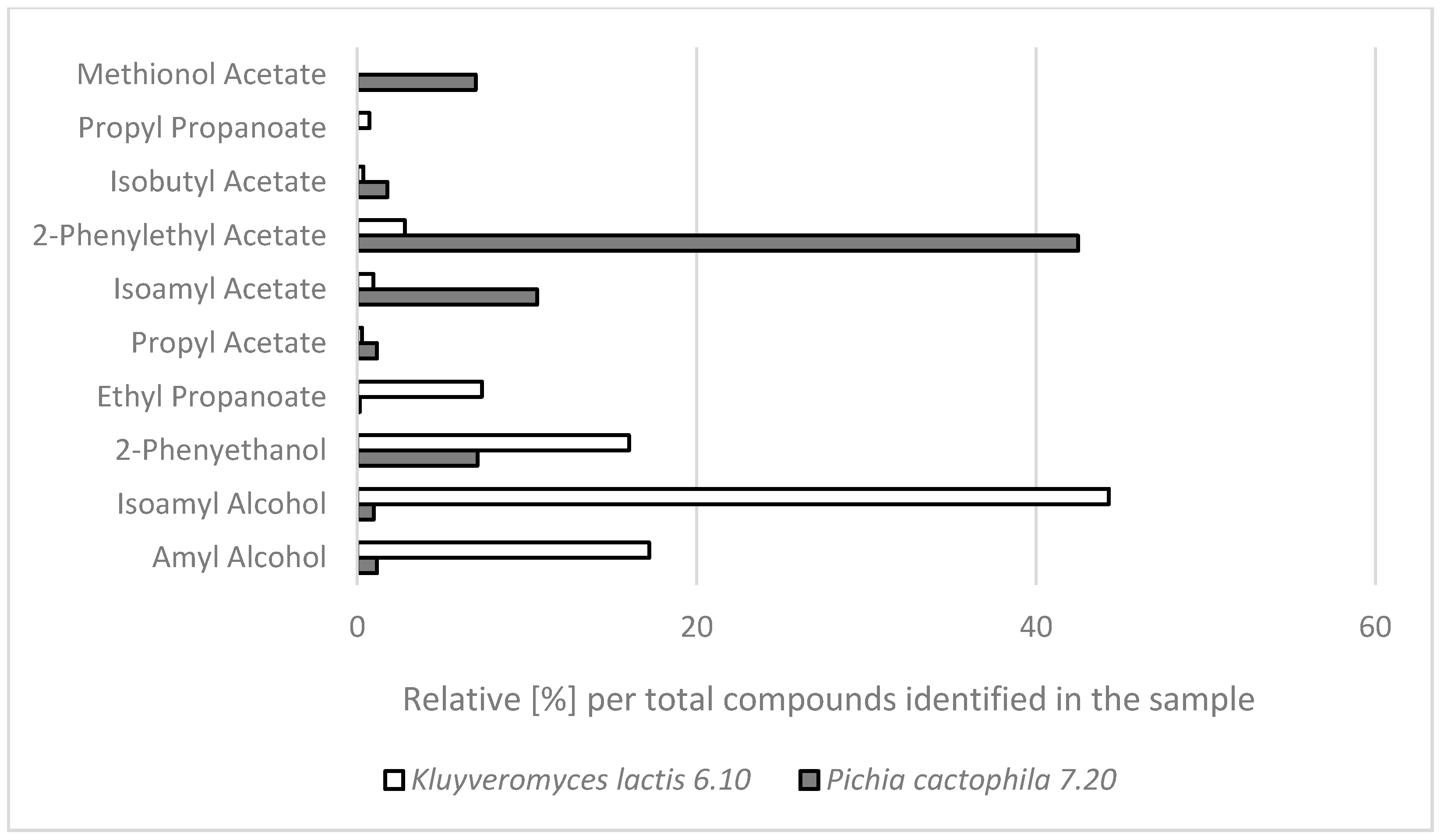
2.2. GC-MS Analysis of Aroma Compounds Produced by the Selected NCY Strains
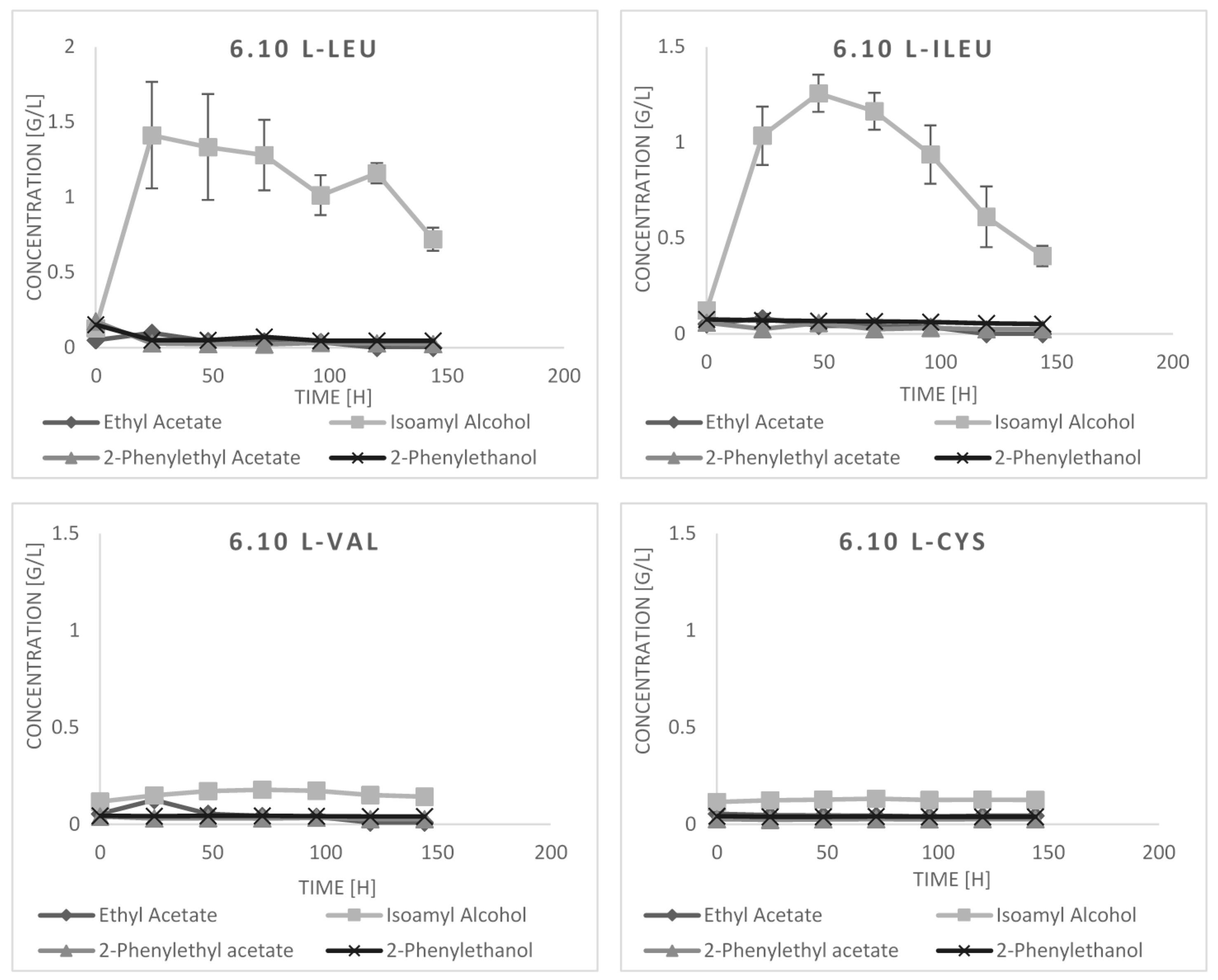
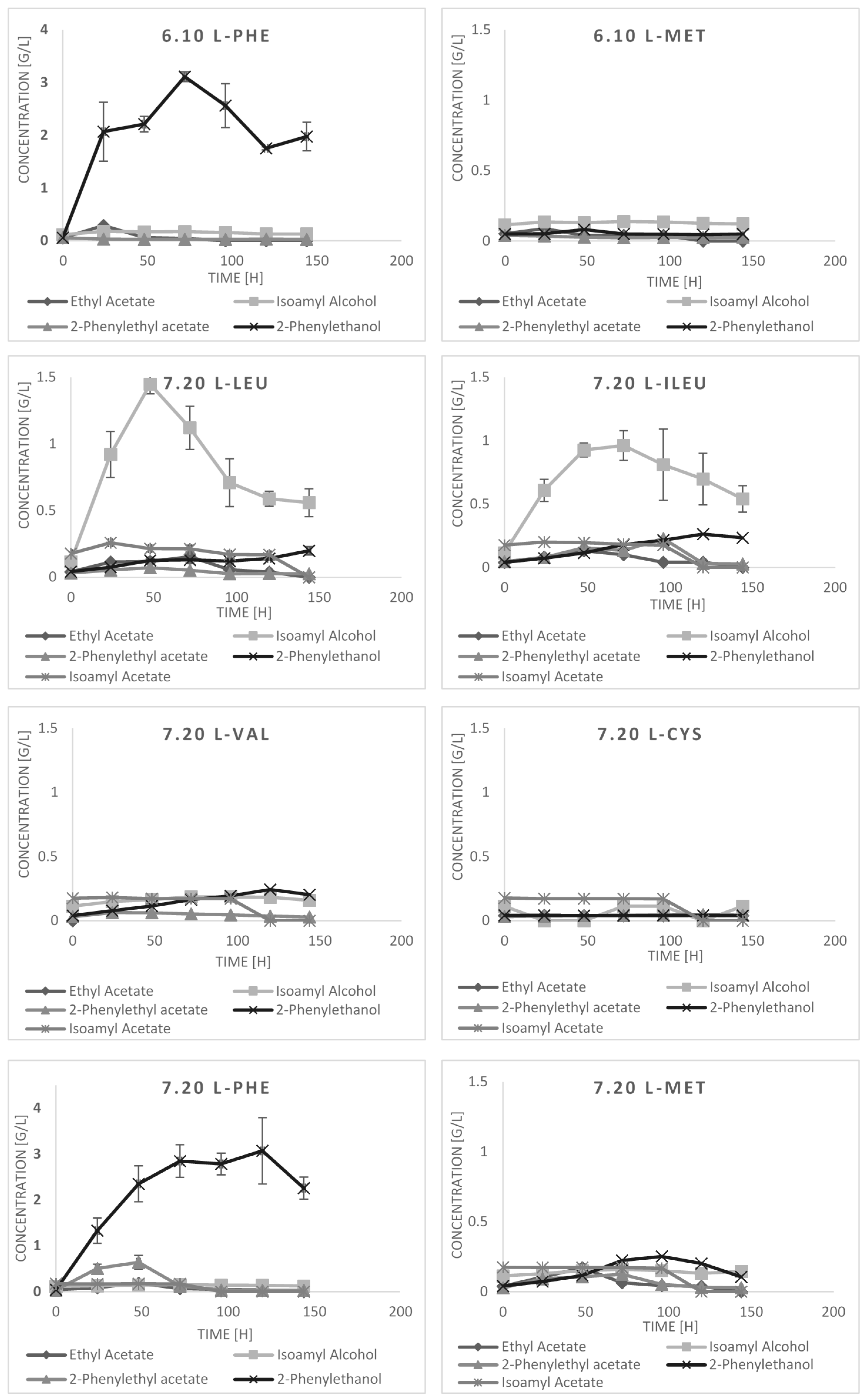
2.3. Bioconversion Cultures
3. Material and Methods
3.1. Environmental Samples and Monocultures Isolation
3.2. Identification of the Isolates—rDNA Region Sequencing
3.3. GC-MS Analysis
3.4. Bioconversion Cultures
3.5. Gas Chromatography
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xu, P.; Hua, D.L.; Ma, C.Q. Microbial transformation of propenylbenzenes for natural flavor production. Trends Biotechnol. 2007, 25, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Forti, L.; Di Mauro, S.; Cramarossa, M.R.; Filippucci, S.; Turchetti, B.; Buzzini, P. Non-Conventional Yeasts Whole Cells as Efficient Biocatalysts for the Production of Flavors and Fragrances. Molecules 2015, 20, 10377–10398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazelwood, L.A.; Daran, J.-M.; van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [PubMed]
- Grondin, E.; Shum, A.; Sing, C.; Caro, Y.; Raherimandimby, M.; Lalaniaina, A.; James, S.; Nueno-palop, C.; Marie, J.; Petit, T. International Journal of Food Microbiology A comparative study on the potential of epiphytic yeasts isolated from tropical fruits to produce fl avoring compounds. Int. J. Food Microbiol. 2015, 203, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Sibirny, A.A.; Scheffers, L. Thematic section: Biochemistry, Genetics, Biotechnology and Ecology of Non-conventional Yeasts. FEMS Yeast Res. 2002, 2, 293. [Google Scholar] [CrossRef]
- Wolf, K. Nonconventional Yeasts in Biotechnology. A Handbook; Springer: Berlin, Germany, 1996. [Google Scholar]
- Etschmann, M.M.; Sell, D.; Schrader, J. Screening of yeasts for the production of the aroma compound 2-phenylethanol in a molasses-based medium. Biotechnol. Lett. 2003, 25, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Sun, B.G.; Cao, Y.P.; Wang, J.; Zhang, H. Biosynthesis of natural 2-phenylethanol by yeast cells. Mod. Chem. Ind. 2008, 28, 38–43. [Google Scholar]
- Wojtatowicz, M.; Chrzanowska, J.; Juszczyk, P.; Skiba, A.; Gdula, A. Identification and biochemical characteristics of yeast microflora of Rokpol cheese. Int. J. Food Microbiol. 2001, 69, 135–140. [Google Scholar] [CrossRef]
- Löser, C.; Urit, T.; Bley, T. Perspectives for the biotechnological production of ethyl acetate by yeasts. Appl. Microbiol. Biotechnol. 2014, 98, 5397–5415. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.P.; Etschmann, M.M.; Schrader, J.; de Billerbeck, G.M. Cell factory applications of the yeast Kluyveromyces marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast 2015, 32, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Etschmann, M.M.; Sell, D.; Schrader, J. Medium optimization for the production of the aroma compound 2-phenylethanol using a genetic algorithm. J. Mol. Catal. B Enzym. 2004, 29, 187–193. [Google Scholar] [CrossRef]
- Garavaglia, J.; Flôres, S.H.; Pizzolato, T.M.; Peralba, M.C.; Ayub, M.A.Z. Bioconversion of L-phenylalanine into 2-phenylethanol by Kluyveromyces marxianus in grape must cultures. World J. Microbiol. Biotechnol. 2007, 23, 1273–1279. [Google Scholar] [CrossRef]
- Schrader, J.; Etschmann, M.M.; Sell, D.; Hilmer, J.M.; Rabenhorst, J. Applied biocatalysis for the synthesis of natural flavour compounds—Current industrial processes and future prospects. Biotechnol. Lett. 2004, 26, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Fuganti, C.; Brenna, E. Biocatalytic preparation of natural flavours and fragrances. Trends Biotechnol. 2005, 23, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweißaufbau der Hefe. Eur. J. Inorg. Chem. 1907, 40, 1027–1047. [Google Scholar] [CrossRef]
- Dickinson, J.R.; Salgado, L.E.; Hewlins, M.J. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 8028–8034. [Google Scholar] [CrossRef] [PubMed]
- Jacques, N.; Casaregola, S. Safety assessment of dairy microorganisms: The hemiascomycetous yeasts. Int. J. Food Microbiol. 2008, 126, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Rossi, J.; Gobetti, M.; Buzzini, P.; Corsetti, A.; Smacchi, E.; De Angelis, M. Yeast in dairy. Ann. Microbiol. Enzym. 1997, 47, 169–183. [Google Scholar]
- Flores, M.; Corral, S.; Cano-García, L.; Salvador, A.; Belloch, C. Yeast strains as potential aroma enhancers in dry fermented sausages. Int. J. Food Microbiol. 2015, 212, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: San Diego, CA, USA, 2011. [Google Scholar]
- Vandamme, E.J.; Soetaert, W. Bioflavours and fragrances via fermentation and biocatalysis. J. Chem. Technol. Biotechnol. 2002, 77, 1323–1332. [Google Scholar] [CrossRef]
- Van Bogaert, I.N.A.; De Maeseneire, S.L.; Develter, D.; Soetaert, W.; Vandamme, E.J. Cloning and characterisation of the glyceraldehyde 3-phosphate dehydrogenase gene of Candida bombicola and use of its promoter. J. Ind. Microbiol. Biotechnol. 2008, 35, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Stribny, J.; Gamero, A.; Pérez-Torrado, R.; Querol, A. Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 2015, 205, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Hua, D.; Xu, P. Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 2011, 29, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Yang, X.; Shen, Q.; Wang, K.; Zhu, T. Optimisation of biotransformation conditions for production of 2-phenylethanol by a Saccharomyces cerevisiae CWY132 mutant. Nat. Prod. Res. 2011, 25, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.R.; Lee, S.L.; Chou, C.C. Production and molar yield of 2-phenylethanol by Pichia fermentans L-5 as affected by some medium components. J. Biosci. Bioeng. 2000, 90, 142–147. [Google Scholar] [CrossRef]
- Lomascolo, A.; Lesage-Meessen, L.; Haon, M.; Navarro, D.; Antona, C.; Faulds, C.; Marcel, A. Evaluation of the potential of Aspergillus niger species for the bioconversion of l-phenylalanine into 2-phenylethanol. World J. Microbiol. Biotechnol. 2001, 17, 99–102. [Google Scholar] [CrossRef]
- Celińska, E.; Kubiak, P.; Białas, W.; Dziadas, M.; Grajek, W. Yarrowia lipolytica: The novel and promising 2-phenylethanol producer. J. Ind. Microbiol. Biotechnol. 2013, 40, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Celińska, E.; Olkowicz, M.; Grajek, W. l-Phenylalanine catabolism and 2-phenylethanol synthesis in Yarrowia lipolytica—Mapping molecular identities through whole-proteome quantitative mass spectrometry analysis. FEMS Yeast Res. 2015, 15, fov041. [Google Scholar] [CrossRef] [PubMed]
- Etschmann, M.M.; Kotter, P.; Hauf, J.; Bluemke, W.; Entian, K.-D.; Schrader, J. Production of the aroma chemicals 3-(methylthio)-1-propanol and 3-(methylthio)-propylacetate with yeasts. Appl. Microbiol. Biotechnol. 2008, 80, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Seow, Y.X.; Ong, P.K.; Liu, S.Q. Production of flavour-active methionol from methionine metabolism by yeasts in coconut cream. Int. J. Food Microbiol. 2010, 143, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Barth, G.; Gaillardin, C. Yarrowia lipolytica. In Nonconventional Yeasts in Biotechnology. A Handbook; Wolf, K., Ed.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1996; pp. 313–388. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 2-phenylethanol, isoamyl alcohol, ethyl acetate, 2-phenylethyl acetate, isoamyl acetate are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celińska, E.; Bonikowski, R.; Białas, W.; Dobrowolska, A.; Słoma, B.; Borkowska, M.; Kubiak, M.; Korpys, P.; Grajek, W. Pichia cactophila and Kluyveromyces lactis are Highly Efficient Microbial Cell Factories of Natural Amino Acid-Derived Aroma Compounds. Molecules 2018, 23, 97. https://doi.org/10.3390/molecules23010097
Celińska E, Bonikowski R, Białas W, Dobrowolska A, Słoma B, Borkowska M, Kubiak M, Korpys P, Grajek W. Pichia cactophila and Kluyveromyces lactis are Highly Efficient Microbial Cell Factories of Natural Amino Acid-Derived Aroma Compounds. Molecules. 2018; 23(1):97. https://doi.org/10.3390/molecules23010097
Chicago/Turabian StyleCelińska, Ewelina, Radosław Bonikowski, Wojciech Białas, Anna Dobrowolska, Barbara Słoma, Monika Borkowska, Monika Kubiak, Paulina Korpys, and Włodzimierz Grajek. 2018. "Pichia cactophila and Kluyveromyces lactis are Highly Efficient Microbial Cell Factories of Natural Amino Acid-Derived Aroma Compounds" Molecules 23, no. 1: 97. https://doi.org/10.3390/molecules23010097




