Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus
Abstract
:1. Introduction
2. Results and Discussion
2.1. Features of A. tataricus cpDNA
2.2. Simple Sequence Repeat (SSR) Analysis
2.3. Comparative Chloroplast Genomic Analysis
2.4. Inverted Repeat (IR) Contraction and Expansion in the A. tataricus Chloroplast Genome
2.5. Phylogenetic Analysis
3. Materials and Methods
3.1. DNA Sequencing, Chloroplast Genome Assembly, and Validation
3.2. Gene Annotation and Sequence Analyses
3.3. Genome Comparison
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, D.X.; Hu, B.Q.; Zhang, M.; Zhang, C.F.; Xu, X.H. Simultaneous separation and determination of phenolic acids, pentapeptides, and triterpenoid saponins in the root of Aster tataricus by high-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2015, 38, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Cheng, S.; Xiang, J.; Yu, B.; Zhang, M.; Zhang, C.; Xu, X. Expectorant, antitussive, anti-inflammatory activities and compositional analysis of Aster tataricus. J. Ethnopharmacol. 2015, 164, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Shan, G.; Qin, G.; Zhen, L.N.; Li, M.H.; Hao, J.L. Advances on chemical components and pharmacological actions of Aster tataricus. Med. Res. Educ. 2012, 29, 73–77. [Google Scholar]
- Zhou, W.B.; Tao, J.Y.; Xu, H.M.; Tan, N.H. Three new antiviral triterpenes from Aster tataricus. Z. Naturforschung B 2010, 65, 1393–1396. [Google Scholar] [CrossRef]
- Tang, X.W.; Liu, X.X.; Tang, Y.L.; Liu, Y.L.; Xu, K.H. Analysis of effective constituents from Aster tataricus L. and extracting of alkaloid and its antibacterial test in vitro. J. Tradit. Chin. Vet. Med. 2006, 1, 16–18. [Google Scholar]
- Du, L.; Mei, H.F.; Yin, X.; Xing, Y.Q. Delayed growth of glioma by a polysaccharide from Aster tataricus, involve upregulation of Bax/Bcl-2 ratio, activation of caspase-3/8/9, and downregulation of the Akt. Tumour Biol. 2014, 35, 1819–1825. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Nagashima, S.; Takeya, K.; Itokawa, H. Solution forms of antitumor cyclic pentapeptides with 3,4-dichlorinated proline residues, astins a and c, from Aster tataricus. Chem. Pharm. Bull. 1995, 43, 1395–1397. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Kai, F.M.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Gene 2008, 9, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Hackett, J.D.; Bhattacharya, D. A genomic and phylogenetic perspective on endosymbiosis and algal origin. J. Appl. Phycol. 2006, 18, 475–481. [Google Scholar] [CrossRef]
- Kanno, A.; Hirai, A. A transcription map of the chloroplast genome from rice (Oryza sativa). Curr. Genet. 1993, 23, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.Z.; Ziersen, B.; Jensen, K.; Lassen, L.M.; Olsen, C.E.; Moller, B.L.; Jensen, P.E. Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth. Biol. 2013, 2, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Echt, C.S.; Deverno, L.L.; Anzidei, M.; Vendramin, G.G. Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol. Ecol. 1998, 7, 307–316. [Google Scholar] [CrossRef]
- Wang, Y.; Ghouri, F.; Shahid, M.Q.; Naeem, M.; Baloch, F.S. The genetic diversity and population structure of wild soybean evaluated by chloroplast and nuclear gene sequences. Biochem. Syst. Ecol. 2017, 71, 170–178. [Google Scholar] [CrossRef]
- Alvespereira, A.; Clement, C.R.; Picanço-Rodrigues, D.; Veasey, E.A.; Dequigiovanni, G.; Ramos, S.L.F.; Pinheiro, J.B.; Zucchi, M.I. Patterns of nuclear and chloroplast genetic diversity and structure of manioc along major Brazilian Amazonian rivers. Ann. Bot. 2018, 121, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Du, L.W.; Liu, A.; Chen, J.J.; Wu, L.; Hu, W.M.; Zhang, W.; Kim, K.H.; Lee, S.C.; Yang, T.J.; et al. The complete chloroplast genome sequences of five epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016, 7, 696. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.B.; Park, H.S.; Lee, S.C.; Lee, J.; Park, J.Y.; Yang, T.J. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J. Agric. Food Chem. 2017, 65, 6298–6306. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huo, N.; Dong, L.; Wang, Y.; Zhang, S.; Young, H.A.; Feng, X.; Gu, Y.Q. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS ONE 2013, 8, e57533. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Khan, M.S.; Allison, L. Milestones in chloroplast genetic engineering: An environmentally friendly era in biotechnology. Trends Plant. Sci. 2002, 7, 84–91. [Google Scholar] [CrossRef]
- Daniell, H.; Kumar, S.; Dufourmantel, N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005, 23, 238–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [PubMed]
- Grosche, C.; Funk, H.T.; Maier, U.G.; Zauner, S. The chloroplast genome of Pellia endiviifolia: Gene content, RNA-editing pattern, and the origin of chloroplast editing. Genome. Biol. Evol. 2012, 4, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.B.; Li, D.Z.; Li, H.T. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Res. 2015, 14, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ma, P.F.; Wen, J.; Yi, T.S. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS ONE 2013, 8, e78568. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Park, S.J. The complete chloroplast genome sequence of Aster spathulifolius (Asteraceae); genomic features and relationship with Asteraceae. Gene 2015, 572, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y.; Yu, Y.; Deng, Y.Q.; Li, J.; Huang, Z.X.; Zhou, S.D. The Chloroplast Genome of Lilium henrici: Genome Structure and Comparative Analysis. Molecules 2018, 23, 1276. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cui, Y.; Chen, X.; Li, Y.; Xu, Z.; Duan, B.; Li, Y.; Song, J.; Yao, H. Complete Chloroplast Genomes of Papaver rhoeas and Papaver orientale: Molecular Structures, Comparative Analysis, and Phylogenetic Analysis. Molecules 2018, 23, 437. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Li, X.; Li, H.; Yang, J.; Wang, H.; He, J. Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species. Molecules 2018, 23, 1015. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, E.; Takahashi, Y.; Lemieux, C.; Turmel, M.; Rochaix, J.D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. Embo. J. 1997, 16, 6095–6104. [Google Scholar] [CrossRef] [PubMed]
- Carbonell-Caballero, J.; Alonso, R.; Ibañez, V.; Terol, J.; Talon, M.; Dopazo, J. A Phylogenetic Analysis of 34 Chloroplast Genomes Elucidates the Relationships between Wild and Domestic Species within the Genus Citrus. Mol. Biol. Evol. 2015, 32, 2015–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zhang, S.; Li, F.; Zhang, S.; Zhang, H.; Wang, X.; Sun, R.; Bonnema, G.; Borm, T.J. A Phylogenetic Analysis of Chloroplast Genomes Elucidates the Relationships of the Six Economically Important Brassica Species Comprising the Triangle of U. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Yang, J. The complete chloroplast genome sequence of Morus mongolica, and a comparative analysis within the Fabidae clade. Curr. Genet. 2016, 62, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. Multiple Independent Losses of Two Genes and One Intron from Legume Chloroplast Genomes. Syst. Bot. 1995, 20, 272–294. [Google Scholar] [CrossRef]
- Nguyen, D.S.; Sai, T.Z.; Nawaz, G.; Lee, K.; Kang, H. Abiotic stresses affect differently the intron splicing and expression of chloroplast genes in coffee plants (Coffea arabica) and rice (Oryza sativa). J. Plant Physiol. 2016, 201, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Mohammad-Panah, N.; Shabanian, N.; Khadivi, A.; Rahmani, M.-S.; Emami, A. Genetic structure of gall oak (Quercus infectoria) characterized by nuclear and chloroplast SSR markers. Tree Genet. Genomes 2017, 13, 70–82. [Google Scholar] [CrossRef]
- Park, S.H.; Sang, I.P.; Gil, J.; Hwangbo, K.; Um, Y.; Kim, H.B.; Jung, C.S.; Kim, S.C.; Lee, Y. Development of Chloroplast SSR Markers to Distinguish Codonopsis Species. Korean Soc. Hortic. Sci. 2017, 5, 207–208. [Google Scholar]
- Zeng, J.; Chen, X.; Wu, X.F.; Jiao, F.C.; Xiao, B.G.; Li, Y.P.; Tong, Z.J. Genetic diversity analysis of genus Nicotiana based on SSR markers in chloroplast genome and mitochondria genome. Acta Tab. Sin. 2016, 22, 89–97. [Google Scholar]
- Park, S.; Sang, I.P.; Gil, J.; Um, Y.; Jung, C.S.; Lee, J.H.; Kim, S.C.; Kim, H.B.; Lee, Y. Development of Simple Sequence Repeat SSR Markers Based on Chloroplast DNA to Distinguish 3 Angelica Species. Korean Soc. Hortic. Sci. 2016, 10, 226. [Google Scholar]
- Zhihai, H.; Jiang, X.; Shuiming, X.; Baosheng, L.; Yuan, G.; Chaochao, Z.; Xiaohui, Q.; Wen, X.; Shilin, C. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. Gigascience 2016, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chu, Y.; Liao, B.; Xiao, S.; Yin, Q.; Bai, R.; Su, H.; Dong, L.; Li, X.; Qian, J.; et al. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience 2017, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kode, V.; Mudd, E.A.; Iamtham, S.; Day, A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. Cell Mol. Biol. 2005, 44, 237–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 2007, 8, 174–201. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The first complete chloroplast genome sequences in actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. Ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Bédard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 2013, 339, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Davis, J.I.; Soreng, R.J.; Garvin, D.; Anderson, M.J. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. USA 1992, 89, 7722–7726. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Palmer, J.D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci. USA 1987, 84, 5818–5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Hahn, F.M.; McMahan, C.M.; Cornish, K.; Whalen, M.C. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009, 9, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Nugent, J.M.; Herbon, L.A. Unusual structure of geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. USA 1987, 84, 769–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raubeson, L.A.; Jansen, R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 1992, 255, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Shams, S.S.; Vahed, S.Z.; Soltanzad, F.; Kafil, V.; Barzegari, A.; Atashpaz, S.; Barar, J. Highly effective DNA extraction method from fresh, frozen, dried and clotted blood samples. Bioimpacts 2011, 1, 183–187. [Google Scholar]
- Simbolo, M.; Gottardi, M.; Corbo, V.; Fassan, M.; Mafficini, A.; Malpeli, G.; Lawlor, R.T.; Scarpa, A. DNA Qualification Workflow for Next Generation Sequencing of Histopathological Samples. PLoS ONE 2013, 8, e62692. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Acemel, R.D.; Tena, J.J.; Irastorzaazcarate, I.; Marlétaz, F.; Gómez-Marín, C.; de la Calle-Mustienes, E.; Bertrand, S.; Diaz, S.G.; Aldea, D.; Aury, J.M.; et al. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 2016, 48, 336–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 2012, 13, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellar Genome DRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, L.; Beszteri, B.; Gäbler-Schwarz, S.; Held, C.; Leese, F.; Mayer, C.; Pöhlmann, K.; Frickenhau, S. STAMP: Extensions to the STADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinform. 2009, 10, 41. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Sequence data of Aster tataricus are available from the authors. |
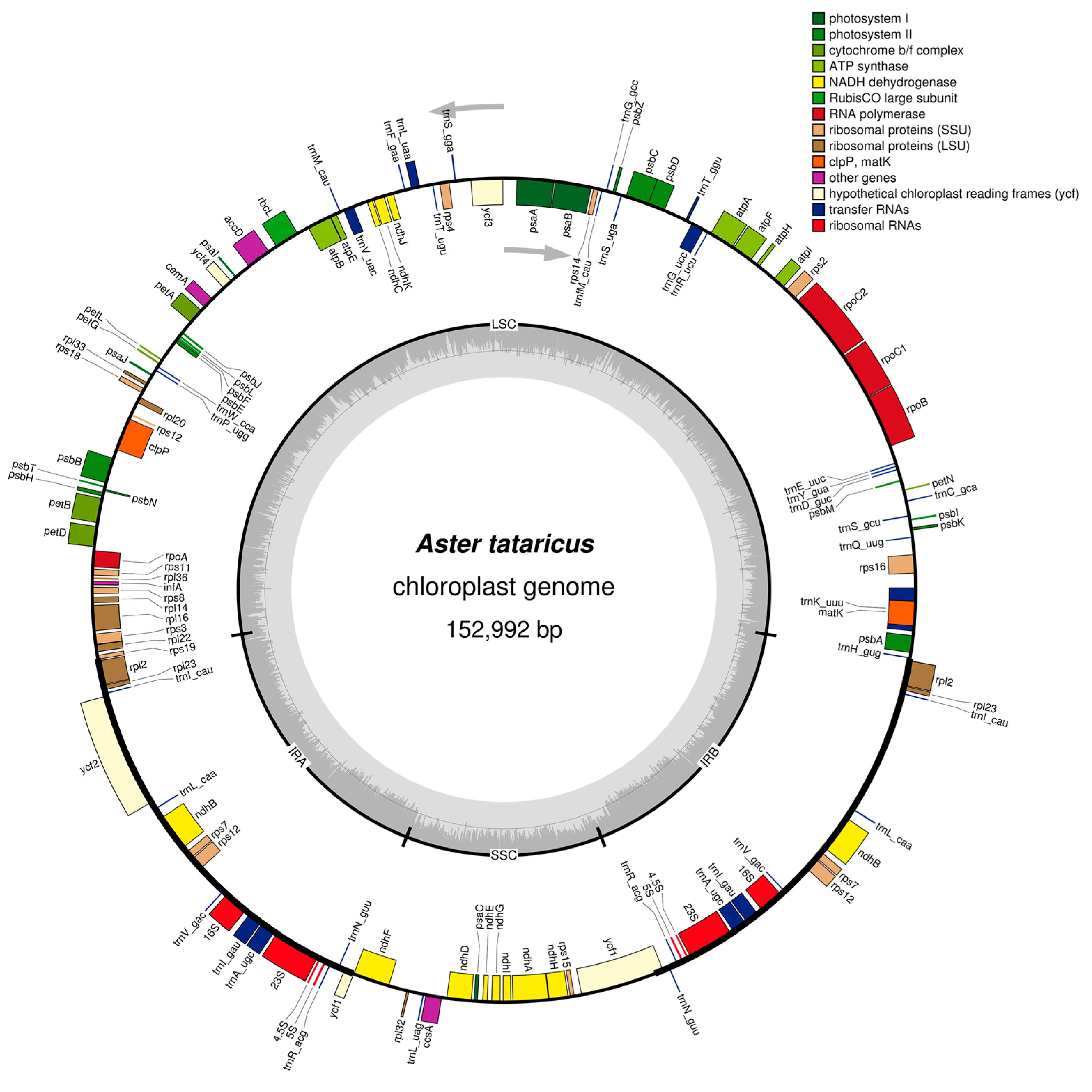
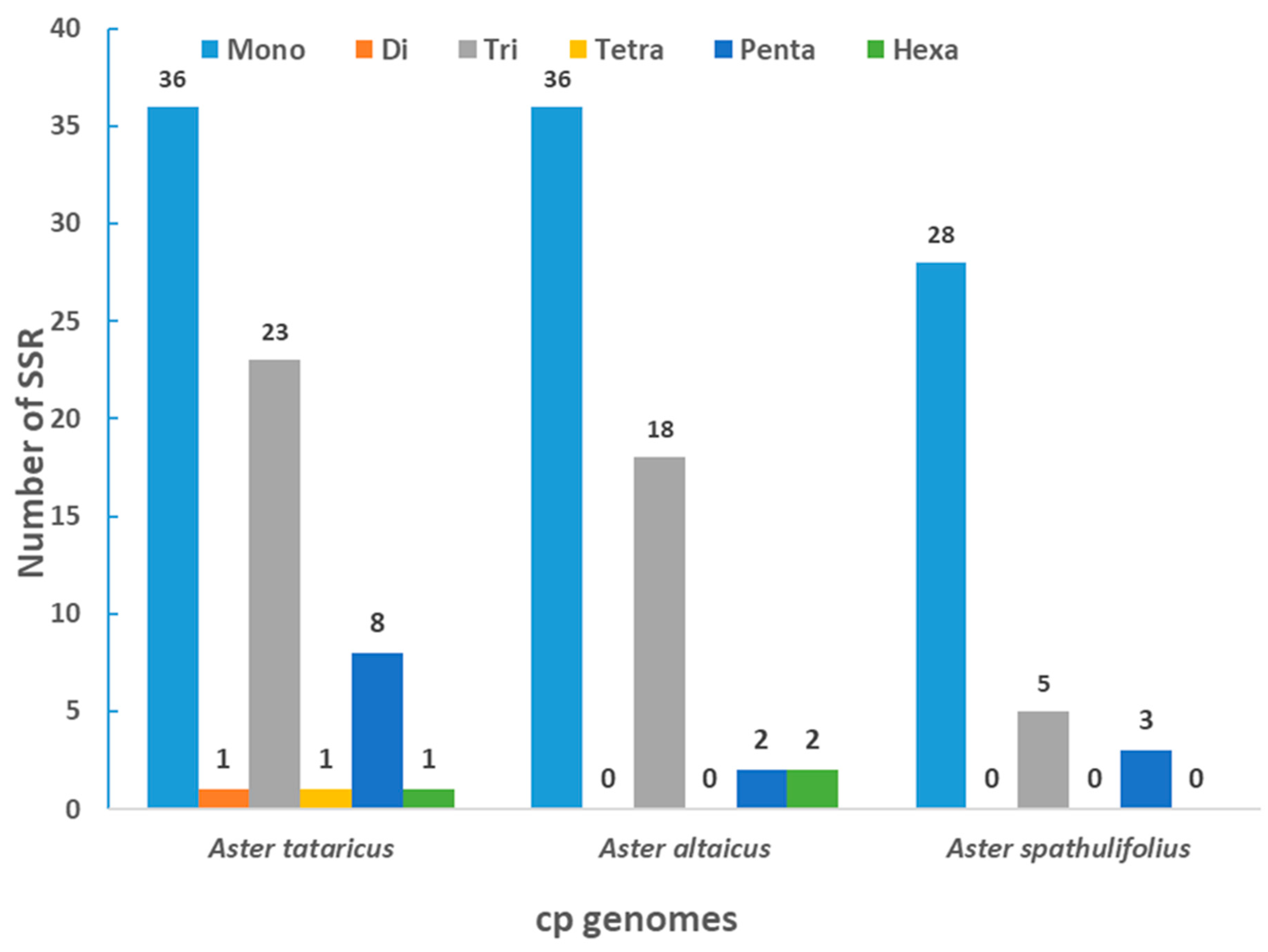


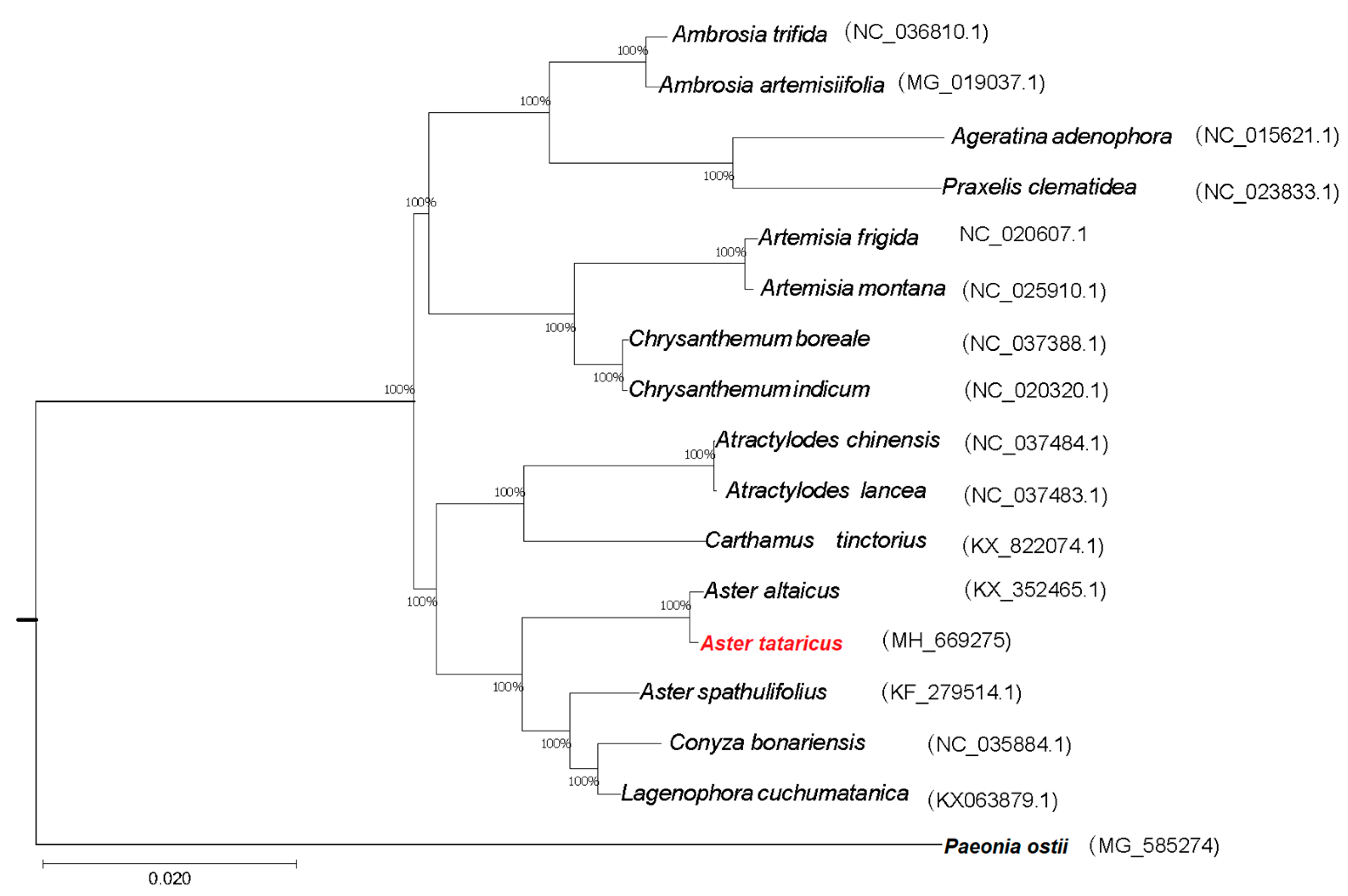
| Species | Aster altaicus | Aster spathulifolius | Aster tataricus |
|---|---|---|---|
| Large single-copy (LSC) | |||
| Length (bp) | 84,240 | 81,998 | 84,698 |
| G + C (%) | 35.3 | 31.4 | 35.2 |
| Length (%) | 55.3 | 54.9 | 55.4 |
| Small single-copy (SSC) | |||
| Length (bp) | 18,196 | 17,973 | 18,250 |
| G + C (%) | 31.3 | 35.8 | 31.3 |
| Length (%) | 11.9 | 12.0 | 11.9 |
| IR | |||
| Length (bp) | 25,005 | 24,751 | 25,022 |
| G + C (%) | 43.0 | 43.2 | 43.0 |
| Length (%) | 16.4 | 16.6 | 16.4 |
| Total | |||
| Length (bp) | 152,446 | 149,473 | 152,992 |
| G + C (%) | 37.3 | 37.7 | 37.3 |
| Category | Gene Group | Gene Names |
|---|---|---|
| Self-replication | Large subunit of ribosomal proteins | rpl2 **,a, 14, 16 **, 20, 22, 23 a, 32, 33, 36 |
| Small subunit of ribosomal proteins | rps2, 3, 4, 7a, 8, 11, 12 **,a, 14, 16 **, 18, 19 | |
| DNA-dependent RNA polymerase | rpoA, B, C1 **, C2 | |
| rRNA genes | rrn16Sa, rrn23Sa, rrn4.5Sa, rrn5Sa | |
| tRNA genes | trnA-UGC **,a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC **, trnG-GCC, trnH-GUG, trnI-CAU, trnI-GAU **,a, trnK-UUU **, trnL-CAA, trnL-UAA **, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC **, trnW-CCA, trnY-GUA | |
| Photosynthesis | Photosystem I | psaA, B, C, I, J |
| Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, | |
| NADH oxidoreductase | ndhA **, B **,a, C, D, E, F, G, H, I, J, K | |
| Cytochrome b6/f complex | petA, B **, D **, G, L, N | |
| ATP synthase | atpA, B, E, F **, H, I | |
| Rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP ** | |
| Envelope membrane protein | cemA | |
| Subunit acetyl-CoA-carboxylase | AccD | |
| c-Type cytochrome synthesis gene | CcsA | |
| Conserved open reading frames | ycf1, 2a, 3 **, 4, 15 |
| Amino Acid | Codon | No. | RSCU * | tRNA | Amino Acid | Codon | No. | RSCU * | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 1064 | 1.34 | Tyr | UAU | 793 | 1.39 | ||
| Phe | UUC | 528 | 0.66 | trnF-GAA | Tyr | UAC | 348 | 0.61 | trnY-GUA |
| Leu | UUA | 554 | 1.55 | trnL-UAA | Stop | UAA | 508 | 1.09 | |
| Leu | UUG | 492 | 1.37 | trnL-CAA | Stop | UAG | 368 | 0.79 | |
| Leu | CUU | 459 | 1.28 | His | CAU | 374 | 1.36 | ||
| Leu | CUC | 205 | 0.57 | His | CAC | 175 | 0.64 | trnH-GUG | |
| Leu | CUA | 272 | 0.76 | trnL-UAG | Gln | CAA | 482 | 1.4 | trnQ-UUG |
| Leu | CUG | 169 | 0.47 | Gln | CAG | 206 | 0.6 | ||
| Ile | AUU | 798 | 1.46 | Asn | AAU | 792 | 1.39 | ||
| Ile | AUC | 397 | 0.73 | trnI-GAU | Asn | AAC | 347 | 0.61 | trnN-GUU |
| Ile | AUA | 441 | 0.81 | trnI-UAU | Lys | AAA | 914 | 1.42 | trnK-UUU |
| Met | AUG | 415 | 1 | trn(f)M-CAU | Lys | AAG | 376 | 0.58 | |
| Val | GUU | 364 | 1.45 | Asp | GAU | 471 | 1.43 | ||
| Val | GUC | 176 | 0.7 | trnV-GAC | Asp | GAC | 187 | 0.57 | trnD-GUC |
| Val | GUA | 318 | 1.26 | trnV-UAC | Glu | GAA | 550 | 1.39 | trnE-UUC |
| Val | GUG | 148 | 0.59 | Glu | GAG | 241 | 0.61 | ||
| Ser | UCU | 509 | 1.37 | Cys | UGU | 345 | 1.15 | ||
| Ser | UCC | 329 | 0.89 | trnS-GGA | Cys | UGC | 253 | 0.85 | trnC-GCA |
| Ser | UCA | 493 | 1.33 | trnS-UGA | Stop | UGA | 524 | 1.12 | |
| Ser | UCG | 292 | 0.79 | Trp | UGG | 491 | 1 | trnW-CCA | |
| Pro | CCU | 259 | 1.29 | Arg | CGU | 204 | 0.73 | trnR-ACG | |
| Pro | CCC | 156 | 0.78 | trnP-GGG | Arg | CGC | 108 | 0.39 | |
| Pro | CCA | 224 | 1.12 | trnP-UGG | Arg | CGA | 282 | 1.01 | |
| Pro | CCG | 164 | 0.82 | Arg | CGG | 173 | 0.62 | ||
| Thr | ACU | 321 | 1.22 | Arg | AGA | 329 | 0.89 | trnR-UCU | |
| Thr | ACC | 246 | 0.93 | trnT-GGU | Arg | AGG | 277 | 0.75 | |
| Thr | ACA | 314 | 1.19 | trnT-UGU | Ser | AGU | 578 | 2.06 | |
| Thr | ACG | 174 | 0.66 | Ser | AGC | 337 | 1.2 | trnS-GCU | |
| Ala | GCU | 250 | 1.25 | Gly | GGU | 315 | 0.95 | ||
| Ala | GCC | 169 | 0.85 | Gly | GGC | 205 | 0.62 | trnG-GCC | |
| Ala | GCA | 242 | 1.21 | trnA-UGC | Gly | GGA | 466 | 1.4 | trnG-UCC |
| Ala | GCG | 138 | 0.69 | Gly | GGG | 342 | 1.03 |
| Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|
| trnK-UUU | LSC | 37 | 2497 | 38 | ||
| 37 | 2502 | 35 | ||||
| trnG-UCC | LSC | 23 | 732 | 48 | ||
| 23 | 723 | 47 | ||||
| trnL-UAA | LSC | 34 | 441 | 50 | ||
| 37 | 423 | 50 | ||||
| trnV-UAC | LSC | 36 | 575 | 37 | ||
| 38 | 573 | 37 | ||||
| trnI-GAU | IR | 38 | 781 | 35 | ||
| 43 | 776 | 35 | ||||
| trnA-UGC | IR | 38 | 820 | 35 | ||
| 38 | 820 | 35 | ||||
| rps12 * | LSC | 234 | 535 | 25 | —— | 114 |
| 114 | —— | 243 | —— | 243 | ||
| rps16 | LSC | 234 | 820 | 40 | ||
| 39 | 826 | 216 | ||||
| rpl16 | LSC | 402 | 1008 | 10 | ||
| —— | —— | —— | ||||
| rpl2 | IR | 391 | 671 | 434 | ||
| 393 | 668 | 435 | ||||
| rpoC1 | LSC | 431 | 709 | 1639 | ||
| 429 | 721 | 1641 | ||||
| ndhA | SSC | 552 | 1055 | 540 | ||
| 553 | 1105 | 540 | ||||
| ndhB | IR | 777 | 674 | 756 | ||
| 777 | 670 | 756 | ||||
| ycf3 | LSC | 124 | 690 | 228 | 739 | 155 |
| 124 | 697 | 230 | 739 | 153 | ||
| petB | LSC | 6 | 754 | 658 | ||
| 6 | 745 | 642 | ||||
| atpF | LSC | 144 | 718 | 411 | ||
| 145 | 699 | 410 | ||||
| clpP | LSC | 71 | 812 | 291 | 614 | 229 |
| 71 | 800 | 291 | 623 | 229 | ||
| petD | LSC | 9 | 645 | 526 | ||
| 9 | 724 | 474 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Guo, S.; Yin, Y.; Zhang, J.; Yin, X.; Liang, C.; Wang, Z.; Huang, B.; Liu, Y.; Xiao, S.; et al. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus. Molecules 2018, 23, 2426. https://doi.org/10.3390/molecules23102426
Shen X, Guo S, Yin Y, Zhang J, Yin X, Liang C, Wang Z, Huang B, Liu Y, Xiao S, et al. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus. Molecules. 2018; 23(10):2426. https://doi.org/10.3390/molecules23102426
Chicago/Turabian StyleShen, Xiaofeng, Shuai Guo, Yu Yin, Jingjing Zhang, Xianmei Yin, Conglian Liang, Zhangwei Wang, Bingfeng Huang, Yanhong Liu, Shuiming Xiao, and et al. 2018. "Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus" Molecules 23, no. 10: 2426. https://doi.org/10.3390/molecules23102426
APA StyleShen, X., Guo, S., Yin, Y., Zhang, J., Yin, X., Liang, C., Wang, Z., Huang, B., Liu, Y., Xiao, S., & Zhu, G. (2018). Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Aster tataricus. Molecules, 23(10), 2426. https://doi.org/10.3390/molecules23102426




