2′-O-Methyl-8-methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA
Abstract
1. Introduction
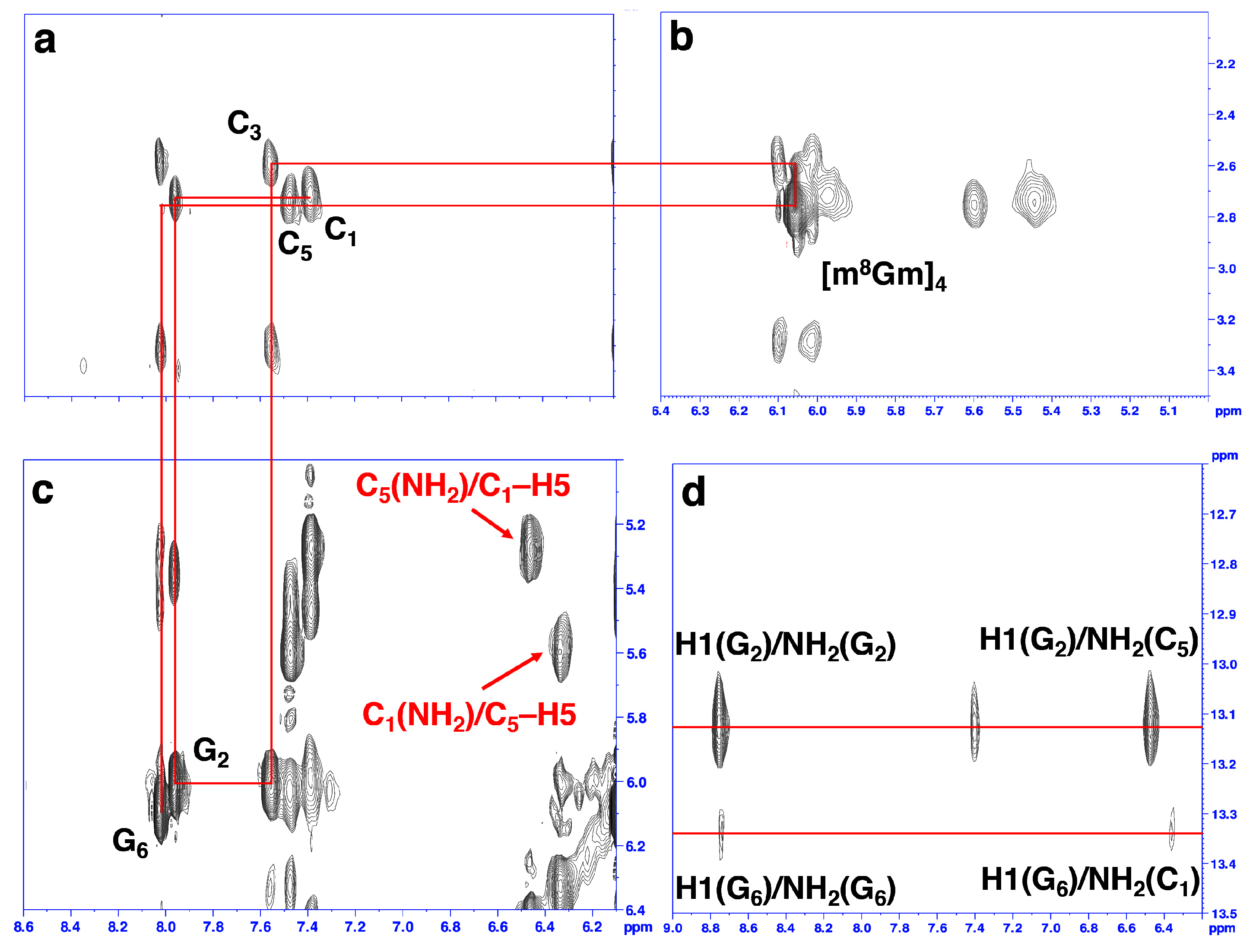
2. Results and Discussion
3. Materials and Methods
3.1. Sample Preparation
3.2. CD Experiments
3.3. NMR Experiments
3.4. Molecular Modeling
3.5. Elctrophoretic Mobility Shift Assay (EMSA)
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Popenda, M.; Biala, E.; Milecki, J.; Adamiak, R.W. Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res. 1997, 25, 4589–4598. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Milecki, J.; Adamiak, R.W. High salt solution structure of a left-handed RNA double helix. Nucleic Acids Res. 2004, 32, 4044–4054. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Rould, M.A.; Lowenhaupt, K.; Herbert, A.; Rich, A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 1999, 284, 1841–1845. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.C.; Lowenhaupt, K.; Rich, A.; Kim, Y.G.; Kim, K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 2005, 437, 1183–1186. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.M.; Bang, J.; Lee, E.H.; Ahn, H.C.; Seo, Y.J.; Kim, K.K.; Kim, Y.G.; Choi, B.S.; Lee, J.H. NMR spectroscopic elucidation of the B-Z transition of a DNA double helix induced by the Z alpha domain of human ADAR1. J. Am. Chem. Soc. 2009, 131, 11485–11491. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, S.H.; Hong, S.C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. USA 2010, 7, 4985–4990. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Kim, D.; Kim, K.K.; Kim, Y.G.; Hohng, S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011, 133, 668–671. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.R.; Park, C.J.; Cheong, H.K.; Ryu, K.S.; Park, J.W.; Kwon, M.Y.; Lee, J.; Kim, K.K.; Choi, B.S.; Lee, J.H. Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition. Nucleic Acids Res. 2016, 44, 2936–2948. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lim, S.H.; Lee, A.R.; Kwon, D.H.; Song, H.K.; Lee, J.H.; Cho, M.; Johner, A.; Lee, N.K.; Hong, S.C. Unveiling the pathway to Z-DNA in the protein-induced B-Z transition. Nucleic Acids Res. 2018, 46, 4129–4137. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Behlke, J.; Lowenhaupt, K.; Heinemann, U.; Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001, 8, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Rothenburg, S.; Deigendesch, N.; Dittmar, K.; Koch-Nolte, F.; Haag, F.; Lowenhaupt, K.; Rich, A. A PKR-like eukaryotic initiation factor 2α kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. USA 2005, 2, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Kahmann, J.D.; Wecking, D.A.; Putter, V.; Lowenhaupt, K.; Kim, Y.G.; Schmieder, P.; Oschkinat, H.; Rich, A.; Schade, M. The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 2712–2717. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H.; Kawai, K.; Matsunaga, A.; Fujimoto, K.; Saito, I.; Robinson, H.; Wang, A.H. Synthesis, structure and thermodynamic properties of 8-methylguanine-containing oligonucleotides: Z-DNA under physiological salt conditions. Nucleic Acids Res. 1996, 24, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Saito, I.; Sugiyama, H. Conformation-Dependent Photochemistry of 5-Halouracil-Containing DNA: Stereospecific 2′α-Hydroxylation of Deoxyribose in Z-form DNA. J. Am. Chem. Soc. 1999, 121, 1391–1392. [Google Scholar] [CrossRef]
- Oyoshi, T.; Kawai, K.; Sugiyama, H. Efficient C2′alpha-hydroxylation of deoxyribose in protein-induced Z-form DNA. J. Am. Chem. Soc. 2003, 125, 1526–1531. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, R.; Sugiyama, H. A nanothermometer based on the different pi stackings of B- and Z-DNA. Angew. Chem. Int. Ed. 2003, 42, 6018–6020. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ikeda, R.; Sugiyama, H. 8-Methylguanosine: A powerful Z-DNA stabilizer. J. Am. Chem. Soc. 2003, 125, 13519–13524. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.X.; Sugiyama, H.; Umano, T.; Osuga, H.; Tanaka, K. (P)-helicene displays chiral selection in binding to Z-DNA. J. Am. Chem. Soc. 2004, 126, 6566–6567. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.Y.; Park, S.; Otomo, H.; Sakashita, S.; Sugiyama, H. Investigation of B-Z transitions with DNA oligonucleotides containing 8-methylguanine. Artif. DNA PNA XNA 2014, 5, e28226. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Otomo, H.; Zheng, L.; Sugiyama, H. Highly emissive deoxyguanosine analogue capable of direct visualization of B-Z transition. Chem. Commun. 2014, 50, 1573–1575. [Google Scholar] [CrossRef] [PubMed]
- Vongsutilers, V.; Gannett, P.M. C8-Guanine modifications: Effect on Z-DNA formation and its role in cancer. Org. Biomol. Chem. 2018, 16, 2198–2209. [Google Scholar] [CrossRef] [PubMed]
- Bothe, J.R.; Lowenhaupt, K.; Al-Hashimi, H.M. Sequence-specific B-DNA flexibility modulates Z-DNA formation. J. Am. Chem. Soc. 2011, 133, 2016–2018. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Lowenhaupt, K.; Wilbert, C.M.; Hanlon, E.B.; Rich, A. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl. Acad. Sci. USA 2000, 97, 13532–13536. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Park, S.; Sugiyama, H. Development of a visible nanothermometer with a highly emissive 2′-O-methylated guanosine analogue. RSC Adv. 2015, 5, 104601–104605. [Google Scholar] [CrossRef]
- Cummins, L.L.; Owens, S.R.; Risen, L.M.; Lesnik, E.A.; Freier, S.M.; McGee, D.; Guinosso, C.J.; Cook, P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995, 23, 2019–2024. [Google Scholar] [CrossRef] [PubMed]
- Uesugi, S.; Ohkubo, M.; Urata, H.; Ikehara, M.; Kobayashi, Y.; Kyogoku, Y. Ribooligonucleotides, r(C-G-C-G) analogs containing 8-substituted guanosine residues, form left-handed duplexes with Z-form-like structure. J. Am. Chem. Soc. 1984, 6, 3675–3676. [Google Scholar] [CrossRef]
- Chen, G.; Znosko, B.M.; Kennedy, S.D.; Krugh, T.R.; Turner, D.H. Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry 2005, 44, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Placido, D.; Brown, B.A., 2nd; Lowenhaupt, K.; Rich, A.; Athanasiadis, A. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure 2007, 15, 395–404. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| Oligonucleotides | NaClO4 (mM) |
|---|---|
| r(CGCGCG)2 | 4090 |
| r(CGC[m8Gm]CG)2 | 880 |
| r(C[m8Gm]C[m8Gm]CG)2 | 100 |
| r(CGCGUGCG)/r(CGCACGCG) | 5430 |
| r(CGCGUGCG)/r(C[m8Gm]CAC[m8Gm]CG) | 2360 |
| r(C[m8Gm]CGU[m8Gm]CG)/r(C[m8Gm]CAC[m8Gm]CG) | 850 |
| Oligonucleotides | Tm1 (°C) | ΔTm (°C) | ∆G298K (kcal/mol) |
|---|---|---|---|
| r(CGCGCG)2 | 33.5 | − | −1.1 |
| r(CGC[m8Gm]CG)2 | 40.0 | +6.5 | −1.7 |
| r(C[m8Gm]C[m8Gm]CG)2 | 45.7 | +12.2 | −2.1 |
| r(CGCGUGCG)/r(CGCACGCG) | 37.7 | − | −1.7 |
| r(CGCGUGCG)/r(C[m8Gm]CAC[m8Gm]CG) | 45.7 | +8.0 | −2.5 |
| r(C[m8Gm]CGU[m8Gm]CG)/r(C[m8Gm]CAC[m8Gm]CG) | 52.7 | +15.0 | −3.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasubramaniyam, T.; Ishizuka, T.; Xiao, C.-D.; Bao, H.-L.; Xu, Y. 2′-O-Methyl-8-methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA. Molecules 2018, 23, 2572. https://doi.org/10.3390/molecules23102572
Balasubramaniyam T, Ishizuka T, Xiao C-D, Bao H-L, Xu Y. 2′-O-Methyl-8-methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA. Molecules. 2018; 23(10):2572. https://doi.org/10.3390/molecules23102572
Chicago/Turabian StyleBalasubramaniyam, Thananjeyan, Takumi Ishizuka, Chao-Da Xiao, Hong-Liang Bao, and Yan Xu. 2018. "2′-O-Methyl-8-methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA" Molecules 23, no. 10: 2572. https://doi.org/10.3390/molecules23102572
APA StyleBalasubramaniyam, T., Ishizuka, T., Xiao, C.-D., Bao, H.-L., & Xu, Y. (2018). 2′-O-Methyl-8-methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA. Molecules, 23(10), 2572. https://doi.org/10.3390/molecules23102572






