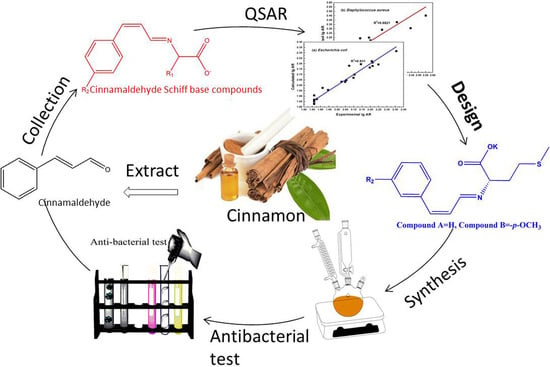
Screening, Synthesis, and QSAR Research on Cinnamaldehyde-Amino Acid Schiff Base Compounds as Antibacterial Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Study on QSAR Models
The Major Structure Descriptors of the Best QSAR Model
2.2. The Synthesized of Screened Compounds
2.3. The Antibacterial Activity of the Screened Compounds
3. Materials and Methods
3.1. Materials
3.2. Method
3.2.1. Determination of Antibacterial Activity
3.2.2. Establishing QSAR Models
3.2.3. Validation of the QSAR Models
3.2.4. Synthesis Methods of Screened Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Barbieri, R.; Coppo, E.; Marchese, A.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Phytochemicals for human disease: An update on plant-derived compounds antibacterial activity. Microchem. Res. 2017, 196, 44–68. [Google Scholar] [CrossRef] [PubMed]
- Marchese, A.; Barbieri, R.; Sanches-Silva, A.; Daglia, M.; Nabavi, S.F.; Jafari, N.J.; Izadi, M.; Ajami, M.; Nabavi, S.M. Antifungal and antibacterial activities of allicin: A review. Trends Food Sci. Technol. 2016, 52, 49–56. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Schiff, H. Mittheilungen aus dem Universitätslaboratorium in Pisa: Eine neue Reihe organischer Basen. Eur. J. Org. Chem. 1864, 131, 118–119. [Google Scholar] [CrossRef]
- Jarrahpour, A.; Khalili, D.; De Clercq, E.; Salmi, C.; Brunel, J.M. Synthesis, antibacterial, antifungal and antiviral activity evaluation of some new bis-Schiff bases of isatin and their derivatives. Molecules 2007, 12, 1720–1730. [Google Scholar] [CrossRef] [PubMed]
- Devi, J.; Devi, S.; Kumar, A. Synthesis, antibacterial evaluation and QSAR analysis of Schiff base complexes derived from [2,2′-(ethylenedioxy)bis(ethylamine)] and aromatic aldehydes. MedChemComm 2016, 7, 932–947. [Google Scholar] [CrossRef]
- Sondhi, S.M.; Arya, S.; Rani, R.; Kumar, N.; Roy, P. Synthesis, anti-inflammatory and anticancer activity evaluation of some mono-and bis-Schiff’s bases. Med. Chem. Res. 2012, 21, 3620–3628. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, X.; Zhao, X.; Meng, R.; Liu, Z.; Chen, X.; Guo, N. Synergistic interactions of nisin in combination with cinnamaldehyde against Staphylococcus aureus in pasteurized milk. Food Control 2017, 71, 10–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Wang, Y.; Jiang, P.; Quek, S. Antibacterial activity and mechanism of cinnamon essential oil against Escherichia coli and Staphylococcus aureus. Food Control 2016, 59, 282–289. [Google Scholar] [CrossRef]
- Moghimi, R.; Aliahmadi, A.; Rafati, H. Ultrasonic nanoemulsification of food grade trans-cinnamaldehyde: 1, 8-Cineol and investigation of the mechanism of antibacterial activity. Ultrason. Sonochem. 2017, 35, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Adabiardakani, A.; Mohammad, H.; Kargar, H. Cinnamaldehyde schiff base derivatives: A short review. World Appl. Program. 2012, 2, 472–476. [Google Scholar]
- Arulmurugan, S.; Kavitha, H.P.; Venkatraman, R. Biological activities of Schiff base and its complexes: A review. Rasayan J. Chem. 2010, 3, 385–410. [Google Scholar]
- Wang, J.; Lian, Z.; Wang, H.; Jin, X.; Liu, Y. Synthesis and antimicrobial activity of Schiff base of chitosan and acylated chitosan. J. Appl. Polym. Sci. 2012, 123, 3242–3247. [Google Scholar] [CrossRef]
- Kudrat-E-Zahan, M.; Islam, M.; Bashar, M.A. Synthesis, characteristics, and antimicrobial activity of some complexes of Mn(II), Fe(III) Co(II), Ni(II), Cu(II), and Sb(III) containing bidentate Schiff base of SMDTC. Russ. J. Gen. Chem. 2015, 85, 667–672. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, H.; Li, S.; Li, Z.; Jiang, M. Synthesis, antimicrobial activity of Schiff base compounds of cinnamaldehyde and amino acids. Bioorg. Med. Chem. Lett. 2016, 26, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Gholivand, K.; EbrahimiValmoozi, A.A.; Gholami, A.; Dusek, M.; Eigner, V.; Abolghasemi, S. Synthesis, characterization, crystal structures, QSAR study and antibacterial activities of organotin bisphosphoramidates. J. Organomet. Chem. 2016, 806, 33–44. [Google Scholar] [CrossRef]
- Verma, J.; Khedkar, V.M.; Coutinho, E.C. 3D-QSAR in drug design-a review. Curr. Top. Med. Chem. 2010, 10, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Fayet, G.; Rotureau, P.; Joubert, L.; Adamo, C. QSPR modeling of thermal stability of nitroaromatic compounds: DFT vs. AM1 calculated descriptors. J. Mol. Model. 2010, 16, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Xiang, J.; Wu, H.; Hu, W.; Zhang, X.; Jin, J.; He, X.; Shen, X.; Zhou, Z.; Fan, S. QSAR modeling of iNOS inhibitors based on a novel regression method: Multi-stage adaptive regression. Chemom. Intell. Lab. Syst. 2013, 128, 83–88. [Google Scholar] [CrossRef]
- Fatemi, M.H.; Ghorbanzad’e, M. In silico prediction of nematic transition temperature for liquid crystals using quantitative structure–property relationship approaches. Mol. Divers. 2009, 13, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Borghini, A.; Pietra, D.; Domenichelli, P.; Bianucci, A.M. QSAR study on thiazole and thiadiazole analogues as antagonists for the adenosine A1 and A3 receptors. Bioorg. Med. Chem. 2005, 13, 5330–5337. [Google Scholar] [CrossRef] [PubMed]
- Eike, D.M.; Brennecke, J.F.; Maginn, E.J. Predicting melting points of quaternary ammonium ionic liquids. Green Chem. 2003, 5, 323–328. [Google Scholar] [CrossRef]
- Jia, L.; Shen, Z.; Su, P. Relationship between reaction rate constants of organic pollutants and their molecular descriptors during Fenton oxidation and in situ formed ferric-oxyhydroxides. J. Environ. Sci. 2016, 43, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Guha, R.; Wild, D.; Chen, T. Ensemble feature selection: Consistent descriptor subsets for multiple QSAR models. J. Chem. Inf. Model. 2007, 47, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chu, W.; Sun, W.; Jiang, C.; Liu, Z. DFT studies of Ni cluster on graphene surface: Effect of CO2 activation. RSC Adv. 2016, 6, 96545–96553. [Google Scholar] [CrossRef]
- Ghasemi, J.; Saaidpour, S. QSPR modeling of stability constants of diverse 15-crown-5 ethers complexes using best multiple linear regression. J. Incl. Phenom. Macrocycl. Chem. 2008, 60, 339–351. [Google Scholar] [CrossRef]
- Beteringhe, A.; Radutiu, A.C.; Culita, D.C.; Mischie, A.; Spafiu, F. Quantitative structure–retention relationship (QSRR) study for predicting gas chromatographic retention times for some stationary phases. Mol. Inf. 2008, 27, 996–1005. [Google Scholar] [CrossRef]
- Lv, P.-C.; Sun, J.; Luo, Y.; Yang, Y.; Zhu, H.-L. Design, synthesis, and structure–activity relationships of pyrazole derivatives as potential FabH inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 4657–4660. [Google Scholar] [CrossRef] [PubMed]
- Stark, P.A.; Abdel-Monem, M. Rumen Protected Essential Amino Acids. U.S. Patent 7,704,521, 27 April 2010. [Google Scholar]
- Yang, D.; Wang, H.; Yuan, H.; Li, S. Quantitative Structure Activity Relationship of Cinnamaldehyde Compounds against Wood-Decaying Fungi. Molecules 2016, 21, 1563. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, H.-J.; Grohe, K. The in vitro and in vivo activity of ciprofloxacin. In Ciprofloxacin; Springer: Berlin, Germany, 1986; pp. 14–18. [Google Scholar] [CrossRef]
- Srivani, P.; Srinivas, E.; Raghu, R.; Sastry, G.N. Molecular modeling studies of pyridopurinone derivatives—Potential phosphodiesterase 5 inhibitors. J. Mol. Graph. Model. 2007, 26, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Sippl, W. Development of biologically active compounds by combining 3D QSAR and structure-based design methods. J. Comput.-Aided Mol. Des. 2002, 16, 825–830. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| ID | Escherichia coli | Staphylococcus aureus | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AR | lgAR | P″, D1 | FPSA3, D2 | ABOC, D3 | RNSB, D4 | AR | lgAR | MASEH, D5 | WNSA1, D6 | RNCS, D7 | NACl, D8 | |
| 1 | 163.64 | 2.2139 | 5.0339 × 10−3 | 0.0098 | 1.0927 | 0.6875 | 91.11 | 1.9596 | −7.2770 | 127.7249 | 13.5586 | 0.0000 |
| 2 | 106.36 | 2.0268 | 2.7696 × 10−3 | 0.0142 | 1.0782 | 0.7222 | 88.89 | 1.9488 | −7.3330 | 126.9269 | 4.5587 | 0.0000 |
| 3 | 157.27 | 2.1967 | 2.1257 × 10−3 | 0.0111 | 1.0915 | 0.6875 | 222.22 | 2.3468 | −7.3530 | 172.4734 | 11.4494 | 1.0000 |
| 4 | 72.73 | 1.8617 | 0.1249 | 0.0111 | 1.1075 | 0.6250 | 88.89 | 1.9488 | −7.3180 | 87.3458 | 10.0842 | 0.0000 |
| 5 | 72.73 | 1.8617 | 2.3127 × 10−3 | 0.0147 | 1.1561 | 0.6786 | 88.89 | 1.9488 | −7.3170 | 90.6090 | 6.6390 | 0.0000 |
| 6 | 163.64 | 2.2139 | 1.6089 × 10−3 | 0.0113 | 1.1059 | 0.6250 | 122.22 | 2.0872 | −7.3100 | 112.3508 | 16.1548 | 1.0000 |
| 7 | 72.73 | 1.8617 | 0.1249 | 0.0120 | 1.0933 | 0.6667 | 96.67 | 1.9853 | −7.3840 | 89.1705 | 8.1225 | 0.0000 |
| 8 | 72.73 | 1.8617 | 2.5943 × 10−3 | 0.0148 | 1.1732 | 0.7097 | 103.33 | 2.0142 | −7.3640 | 119.4079 | 6.6248 | 0.0000 |
| 9 | 113.64 | 2.0555 | 1.7586 × 10−3 | 0.0112 | 1.1650 | 0.6667 | 355.56 | 2.5509 | −7.3800 | 138.2133 | 16.1512 | 1.0000 |
| 10 | 75.45 | 1.8777 | 0.1249 | 0.0106 | 1.0584 | 0.7692 | 88.89 | 1.9488 | −7.3180 | 89.6997 | 12.0427 | 0.0000 |
| 11 | 127.27 | 2.1047 | 2.8601 × 10−3 | 0.0127 | 1.0487 | 0.7907 | 88.89 | 1.9488 | −7.3340 | 110.4771 | 5.3309 | 0.0000 |
| 12 | 161.82 | 2.2090 | 1.9326 × 10−3 | 0.0097 | 1.0579 | 0.7692 | 141.11 | 2.1496 | −7.3170 | 132.7288 | 12.8490 | 1.0000 |
| 13 | 72.73 | 1.8617 | 0.1250 | 0.0117 | 1.0725 | 0.7273 | 88.89 | 1.9488 | −7.3350 | 108.6156 | 5.7232 | 0.0000 |
| 14 | 88.18 | 1.9454 | 3.0117 × 10−3 | 0.0140 | 1.1118 | 0.7568 | 88.89 | 1.9488 | −7.3360 | 115.2242 | 5.8626 | 0.0000 |
| 15 | 139.09 | 2.1433 | 2.0303 × 10−3 | 0.0106 | 1.0716 | 0.7273 | 174.44 | 2.2417 | −7.3300 | 139.9473 | 14.6326 | 1.0000 |
| 16 | 177.27 | 2.2486 | 4.2908 × 10−3 | 0.0107 | 1.1169 | 0.6053 | 127.78 | 2.1065 | −7.3210 | 134.7791 | 13.4207 | 0.0000 |
| 17 | 111.82 | 2.0485 | 3.8575 × 10−3 | 0.0136 | 1.1250 | 0.6429 | 103.33 | 2.0142 | −7.3480 | 157.7156 | 3.6624 | 0.0000 |
| 18 | 118.18 | 2.0726 | 4.2916 × 10−3 | 0.0110 | 1.1164 | 0.6053 | 144.44 | 2.1597 | −7.3080 | 172.0321 | 7.1417 | 1.0000 |
| 19 | 81.82 | 1.9128 | 0.1251 | 0.0112 | 1.0518 | 0.6053 | 88.89 | 1.9488 | −7.3450 | 111.6619 | 6.6481 | 0.0000 |
| 20 | 125.45 | 2.0985 | 2.6664 × 10−3 | 0.0135 | 1.1063 | 0.6429 | 92.22 | 1.9648 | −7.2920 | 175.5388 | 4.9846 | 0.0000 |
| 21 | 227.27 | 2.3565 | 1.9517 × 10−3 | 0.0106 | 1.0512 | 0.6053 | 277.78 | 2.4437 | −7.3320 | 154.3464 | 13.3231 | 1.0000 |
| No | X | ΔX | t test Value | Name of Descriptor |
|---|---|---|---|---|
| E. coli model: R2 = 0.9354, F = 57.96, and s2 = 0.0020 | ||||
| 0 | 5.0709 | 4.0665 × 10−1 | 14.8035 | Intercept |
| 1 | −2.5685 | 1.9802 × 10−1 | −13.5819 | Polarity parameter/square distance, D1 |
| 2 | −4.1057 × 10 | 7.4151 | −5.5369 | FPSA3 Fractional PPSA (PPSA-3/TMSA) [Zefirov’s PC], D2 |
| 3 | −1.7850 | 3.4648 × 10−1 | −5.1519 | Avg bond order of a C atom, D3 |
| 4 | −7.2082 × 10−1 | 1.8119 × 10−1 | −3.9783 | Relative number of single bonds, D4 |
| S. aureus model: R2 = 0.8946, F = 33.94, and s2 = 0.0043 | ||||
| 0 | −1.8664 × 10 | 4.3463 | −4.2942 | Intercept |
| 1 | −2.7525 | 5.8814 × 10−1 | −4.6799 | Min atomic state energy for an H atom, D5 |
| 2 | 2.6523 × 10−3 | 6.9487 × 10−4 | 3.8170 | WNSA-1 Weighted PNSA(PNSA1 × TMSA/100)[Quantum-Chemical PC], D6 |
| 3 | 1.9642 × 10−2 | 5.2856 × 10−3 | 3.7162 | RNCS Relative negative charged SA(SAMNEG × RNCG)[Zefirov’s PC], D7 |
| 4 | 1.1873 × 10−1 | 5.2091 × 10−2 | 2.2794 | Number of Cl atoms, D8 |
| Training Set | N | R2 (fit) | F (fit) | s2 (fit) | Test Set | N | R2 (pred) | F (pred) | s2 (pred) |
|---|---|---|---|---|---|---|---|---|---|
| Validation for the model of E. coli | |||||||||
| A | 14 | 0.9456 | 39.08 | 0.0020 | c | 7 | 0.8759 | 42.36 | 0.0171 |
| B | 14 | 0.8951 | 19.20 | 0.0030 | b | 7 | 0.9826 | 338.07 | 0.0096 |
| C | 14 | 0.9816 | 120.20 | 0.0007 | a | 7 | 0.7800 | 21.27 | 0.0227 |
| Average | 14 | 0.9408 | 59.49 | 0.0019 | Average | 7 | 0.8795 | 133.9 | 0.0165 |
| D | 16 | 0.9262 | 34.52 | 0.0023 | d | 5 | 0.9620 | 101.25 | 0.0162 |
| Validation for the model of S. aureus | |||||||||
| A | 14 | 0.9339 | 31.79 | 0.0037 | c | 7 | 0.6790 | 12.69 | 0.0322 |
| B | 14 | 0.9121 | 23.35 | 0.0044 | b | 7 | 0.8410 | 31.75 | 0.0273 |
| C | 14 | 0.8754 | 15.81 | 0.0046 | a | 7 | 0.9089 | 59.84 | 0.0259 |
| Average | 14 | 0.9071 | 23.65 | 0.0042 | Average | 7 | 0.8096 | 34.76 | 0.0282 |
| D | 16 | 0.8975 | 24.08 | 0.0056 | d | 5 | 0.8029 | 16.29 | 0.0175 |
| No. | Staphylococcus aureus | Escherichia coli | ||||||
|---|---|---|---|---|---|---|---|---|
| Exp.AR | Exp.lgAR | Cal.lgAR | Error | Exp.AR | Exp.lgAR | Cal.lgAR | Error | |
| A | 133.33 | 2.1249 | 1.8365 | 0.2884 | 136.36 | 2.1347 | 1.8831 | 0.2516 |
| B | 159.22 | 2.2020 | 2.0876 | 0.1144 | 136.36 | 2.1347 | 1.9323 | 0.2024 |
| C | 203.33 | 2.3082 | 2.3131 | −0.0049 | 109.09 | 2.0378 | 2.1099 | −0.0721 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Jiang, M.; Sun, F.; Li, S.; Hse, C.-Y.; Jin, C. Screening, Synthesis, and QSAR Research on Cinnamaldehyde-Amino Acid Schiff Base Compounds as Antibacterial Agents. Molecules 2018, 23, 3027. https://doi.org/10.3390/molecules23113027
Wang H, Jiang M, Sun F, Li S, Hse C-Y, Jin C. Screening, Synthesis, and QSAR Research on Cinnamaldehyde-Amino Acid Schiff Base Compounds as Antibacterial Agents. Molecules. 2018; 23(11):3027. https://doi.org/10.3390/molecules23113027
Chicago/Turabian StyleWang, Hui, Mingyue Jiang, Fangli Sun, Shujun Li, Chung-Yun Hse, and Chunde Jin. 2018. "Screening, Synthesis, and QSAR Research on Cinnamaldehyde-Amino Acid Schiff Base Compounds as Antibacterial Agents" Molecules 23, no. 11: 3027. https://doi.org/10.3390/molecules23113027