Demethoxycurcumin-Loaded Chitosan Nanoparticle Downregulates DNA Repair Pathway to Improve Cisplatin-Induced Apoptosis in Non-Small Cell Lung Cancer
Abstract
:1. Introduction
2. Results
2.1. Characterization of the Characteristics of Unloaded CHC Nanoparticles (such as Hydrodynamic Radius (Dh), Zeta-Potential, TEM/SEM Morphology), and to Compare with that of DMC-CHC Nanoparticles (DMC-CHC NPs)
2.2. In Vitro Cytotoxicity
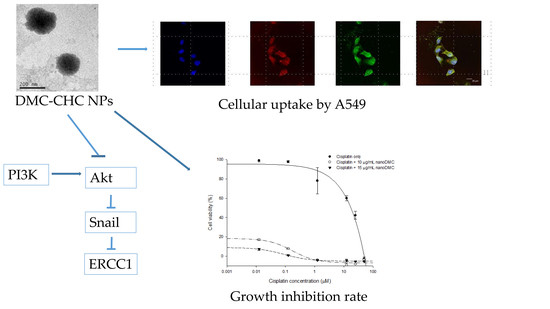
2.3. Fluorescence Confocal Microscopy
2.4. Cellular Uptake
2.5. DMC-CHC NPs Further Decreased Cisplatin-Induced Cell Viability
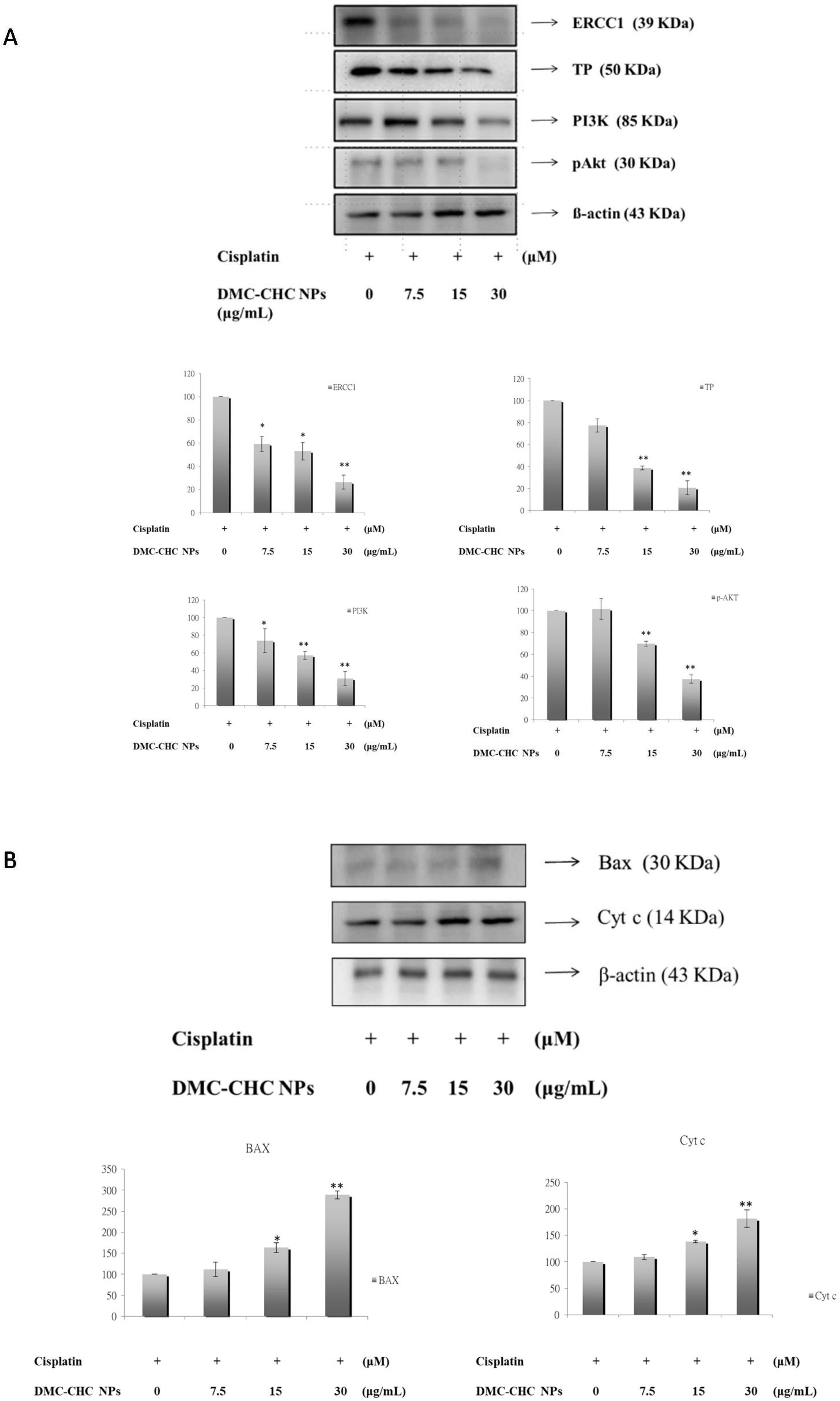
2.6. DMC-CHC NPs Reduced TP and ERCC1 Protein Expressions in Cisplatin-Treated Cell
2.7. DMC-CHC NPs Decreased Protein Levels of PI3K, and Phosphorylation AKT Induced by Cisplatin
2.8. DMC-CHC NPs Altered Protein Levels of Bax, and Cyt c Induced by Cisplatin
3. Discussion
4. Materials and Methods
4.1. Agents
4.2. Characterization of CHC and Preparation of DMC-CHC NPs
4.3. TEM Image
4.4. Determination of the Loading Efficiency and the Drug Encapsulation Efficiency
4.5. Cells and Cell Culture
4.6. Fluorescence Confocal Microscopy
4.7. Cellular Uptake
4.8. Cell Viability Assay (MTT Assay)
4.9. Sulforhodamine B Assay
4.10. Western Blotting Analysis
4.11. Statistics Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tsvetkova, E.; Goss, G. Drug resistance and its significance for treatment decisions in non-small-cell lung cancer. Curr. Oncol. 2012, 19, S45. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-H.; Hua, D.; Du, X. Polymorphisms in p53, GSTP1 and XRCC1 predict relapse and survival of gastric cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Cancer Chemother. Pharmacol. 2009, 64, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nature Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef]
- Mu, H.; Geacintov, N.E.; Broyde, S.; Yeo, J.-E.; Schärer, O.D. Molecular basis for damage recognition and verification by XPC-RAD23B and TFIIH in nucleotide excision repair. DNA Repair 2018, 71, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Olaussen, K.A.; Dunant, A.; Fouret, P.; Brambilla, E.; André, F.; Haddad, V.; Taranchon, E.; Filipits, M.; Pirker, R.; Popper, H.H. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. New Eng. J. Med. 2006, 355, 983–991. [Google Scholar] [CrossRef] [PubMed]
- McNeil, E.M.; Melton, D.W. DNA repair endonuclease ERCC1–XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucl.Acids Res. 2012, 40, 9990–10004. [Google Scholar] [CrossRef]
- Bera, H.; Chigurupati, S. Recent discovery of non-nucleobase thymidine phosphorylase inhibitors targeting cancer. Eur. J. Med. Chem. 2016, 124, 992–1003. [Google Scholar] [CrossRef]
- Elamin, Y.Y.; Rafee, S.; Osman, N.; Kenneth, J.; Gately, K. Thymidine phosphorylase in cancer; enemy or friend? Cancer Microenv. 2016, 9, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Kobayashi, H.; Suzuki, M.; Kanayama, N.; Suzuki, M.; Terao, T. Thymidine phosphorylase expression in tumor-infiltrating macrophages may be correlated with poor prognosis in uterine endometrial cancer. Human Pathol. 2002, 33, 1105–1113. [Google Scholar] [CrossRef]
- Chen, H.W.; Huang, H.C. Effect of curcumin on cell cycle progression and apoptosis in vascular smooth muscle cells. Br. J. Pharm. 1998, 124, 1029–1040. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.-S.; Weng, S.-H.; Kuo, Y.-H.; Chiu, Y.-F.; Lin, Y.-W. Synergistic effect of curcumin and cisplatin via down-regulation of thymidine phosphorylase and excision repair cross-complementary 1 (ERCC1). Mol. Pharm. 2011, 80, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Hung, C.-C.; Wang, C.C.; Lin, H.-Y.; Huang, S.-H.; Sheu, M.-J. Demethoxycurcumin sensitizes the response of non-small cell lung cancer to cisplatin through downregulation of TP and ERCC1-related pathways. Phytomedicine 2018, 53, 28–36. [Google Scholar] [CrossRef]
- Datta, S.; Misra, S.K.; Saha, M.L.; Lahiri, N.; Louie, J.; Pan, D.; Stang, P.J. Orthogonal self-assembly of an organoplatinum (II) metallacycle and cucurbit [8] uril that delivers curcumin to cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, 8087–8092. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Chen, S.-Y.; Liu, D.-M.; Liu, T.-Y. Self-assembled hollow nanocapsule from amphiphatic carboxymethyl-hexanoyl chitosan as drug carrier. Macromolecules 2008, 41, 6511–6516. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Lin, H.-Y.; Wu, C.-H.; Liu, D.-M. Forming of demethoxycurcumin nanocrystallite-chitosan nanocarrier for controlled low dose cellular release for inhibition of the migration of vascular smooth muscle cells. Mol. Pharm. 2012, 9, 2268–2279. [Google Scholar] [CrossRef]
- Huang, W.-T.; Larsson, M.; Wang, Y.-J.; Chiou, S.-H.; Lin, H.-Y.; Liu, D.-M. Demethoxycurcumin-carrying chitosan–antibody core–shell nanoparticles with multitherapeutic efficacy toward malignant A549 lung tumor: From in vitro characterization to in vivo evaluation. Mol. Pharm. 2015, 12, 1242–1249. [Google Scholar] [CrossRef]
- Huang, W.-T.; Larsson, M.; Lee, Y.-C.; Liu, D.-M.; Chiou, G.-Y. Dual drug-loaded biofunctionalized amphiphilic chitosan nanoparticles: Enhanced synergy between cisplatin and demethoxycurcumin against multidrug-resistant stem-like lung cancer cells. Eur. J. Pharm. Biopharm. 2016, 109, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, M.-H.; Larsson, M.; Larsson, A.; Evenbratt, H.; Chen, Y.-Y.; Chen, Y.-Y.; Liu, D.-M. Design and characterization of a novel amphiphilic chitosan nanocapsule-based thermo-gelling biogel with sustained in vivo release of the hydrophilic anti-epilepsy drug ethosuximide. J. Controlled Release 2012, 161, 942–948. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-J.; Chien, Y.-C.; Wu, C.-H.; Liu, D.-M. Magnolol-loaded core–shell hydrogel nanoparticles: Drug release, intracellular uptake, and controlled cytotoxicity for the inhibition of migration of vascular smooth muscle cells. Mol. Pharm. 2011, 8, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Iversen, T.-G.; Skotland, T.; Sandvig, K. Endocytosis and intracellular transport of nanoparticles: Present knowledge and need for future studies. Nano Today 2011, 6, 176–185. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Chan, W.C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, K.; Wang, X.; Chen, H.; Zhou, J.; Wu, X.; Liu, T.; Yang, Y.; Yang, X.; Cui, D. NDRG1 disruption alleviates cisplatin/sodium glycididazole-induced DNA damage response and apoptosis in ERCC1-defective lung cancer cells. Int. J. Biochem. Cell Biol. 2018, 100, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Chirnomas, D.; Taniguchi, T.; de la Vega, M.; Vaidya, A.P.; Vasserman, M.; Hartman, A.-R.; Kennedy, R.; Foster, R.; Mahoney, J.; Seiden, M.V. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol. Cancer Ther. 2006, 5, 952–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Grady, S.; Finn, S.P.; Cuffe, S.; Richard, D.J.; O’Byrne, K.J.; Barr, M.P. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat. Rev. 2014, 40, 1161–1170. [Google Scholar] [CrossRef]
- Roberts, C.M.; Tran, M.A.; Pitruzzello, M.C.; Wen, W.; Loeza, J.; Dellinger, T.H.; Mor, G.; Glackin, C.A. TWIST1 drives cisplatin resistance and cell survival in an ovarian cancer model, via upregulation of GAS6, L1CAM, and Akt signalling. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Krishnamurthy, S. Cellular responses to cisplatin-induced dna damage. J. Nucleic Acids 2010, 2010, 1–16. [Google Scholar] [CrossRef]
- Hung, C.-M.; Su, Y.-H.; Lin, H.-Y.; Lin, J.-N.; Liu, L.-C.; Ho, C.-T.; Way, T.-D. Demethoxycurcumin modulates prostate cancer cell proliferation via AMPK-induced down-regulation of HSP70 and EGFR. J. Agric. Food Chem. 2012, 60, 8427–8434. [Google Scholar] [CrossRef]
- Sheu, M.J.; Lin, H.Y.; Yang, Y.H.; Chou, C.J.; Chien, Y.C.; Wu, T.S.; Wu, C.H. Demethoxycurcumin, a major active curcuminoid from Curcuma longa, suppresses balloon injury induced vascular smooth muscle cell migration and neointima formation: An in vitro and in vivo study. Mol. Nutr. Food Res. 2013, 57, 1586–1597. [Google Scholar] [CrossRef]
- Teng, Y.-N.; Hsieh, Y.-W.; Hung, C.-C.; Lin, H.-Y. Demethoxycurcumin modulates human P-glycoprotein function via uncompetitive inhibition of ATPase hydrolysis activity. J. Agric. Food Chem. 2015, 63, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-T.; Ko, Y.-P.; Kuo, T.-Y.; Larsson, M.; Chang, M.-C.; Jean, R.-D.; Liu, D.-M. A new type of gadodiamide-conjugated amphiphilic chitosan nanoparticle and its use for MR imaging with significantly enhanced contrastability. Carbohydr. Polym. 2019, 203, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Li, P.-C.; Pan, C.-H.; Sheu, M.-J.; Wu, C.-C.; Ma, W.-F.; Wu, C.-H. Deep sea water prevents balloon angioplasty-induced hyperplasia through MMP-2: An in vitro and in vivo study. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds DMC, DMC-CHC NPs are available from the authors. |



| Sample | Dh (nm) | Zeta Potential (mV) |
|---|---|---|
| CHC NPs | 50 (10) | −22 (0.65) |
| DMC-CHC NPs | 130 (21) | −9 (1.1) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-Y.; Lin, Y.-J.; Huang, W.-T.; Hung, C.-C.; Lin, H.-Y.; Tu, Y.-C.; Liu, D.-M.; Lan, S.-J.; Sheu, M.-J. Demethoxycurcumin-Loaded Chitosan Nanoparticle Downregulates DNA Repair Pathway to Improve Cisplatin-Induced Apoptosis in Non-Small Cell Lung Cancer. Molecules 2018, 23, 3217. https://doi.org/10.3390/molecules23123217
Chen Y-Y, Lin Y-J, Huang W-T, Hung C-C, Lin H-Y, Tu Y-C, Liu D-M, Lan S-J, Sheu M-J. Demethoxycurcumin-Loaded Chitosan Nanoparticle Downregulates DNA Repair Pathway to Improve Cisplatin-Induced Apoptosis in Non-Small Cell Lung Cancer. Molecules. 2018; 23(12):3217. https://doi.org/10.3390/molecules23123217
Chicago/Turabian StyleChen, Ying-Yi, Yu-Jung Lin, Wei-Ting Huang, Chin-Chuan Hung, Hui-Yi Lin, Yu-Chen Tu, Dean-Mo Liu, Shou-Jen Lan, and Ming-Jyh Sheu. 2018. "Demethoxycurcumin-Loaded Chitosan Nanoparticle Downregulates DNA Repair Pathway to Improve Cisplatin-Induced Apoptosis in Non-Small Cell Lung Cancer" Molecules 23, no. 12: 3217. https://doi.org/10.3390/molecules23123217