Drug Repurposing for Japanese Encephalitis Virus Infection by Systems Biology Methods
Abstract
1. Introduction
2. Results and Discussion
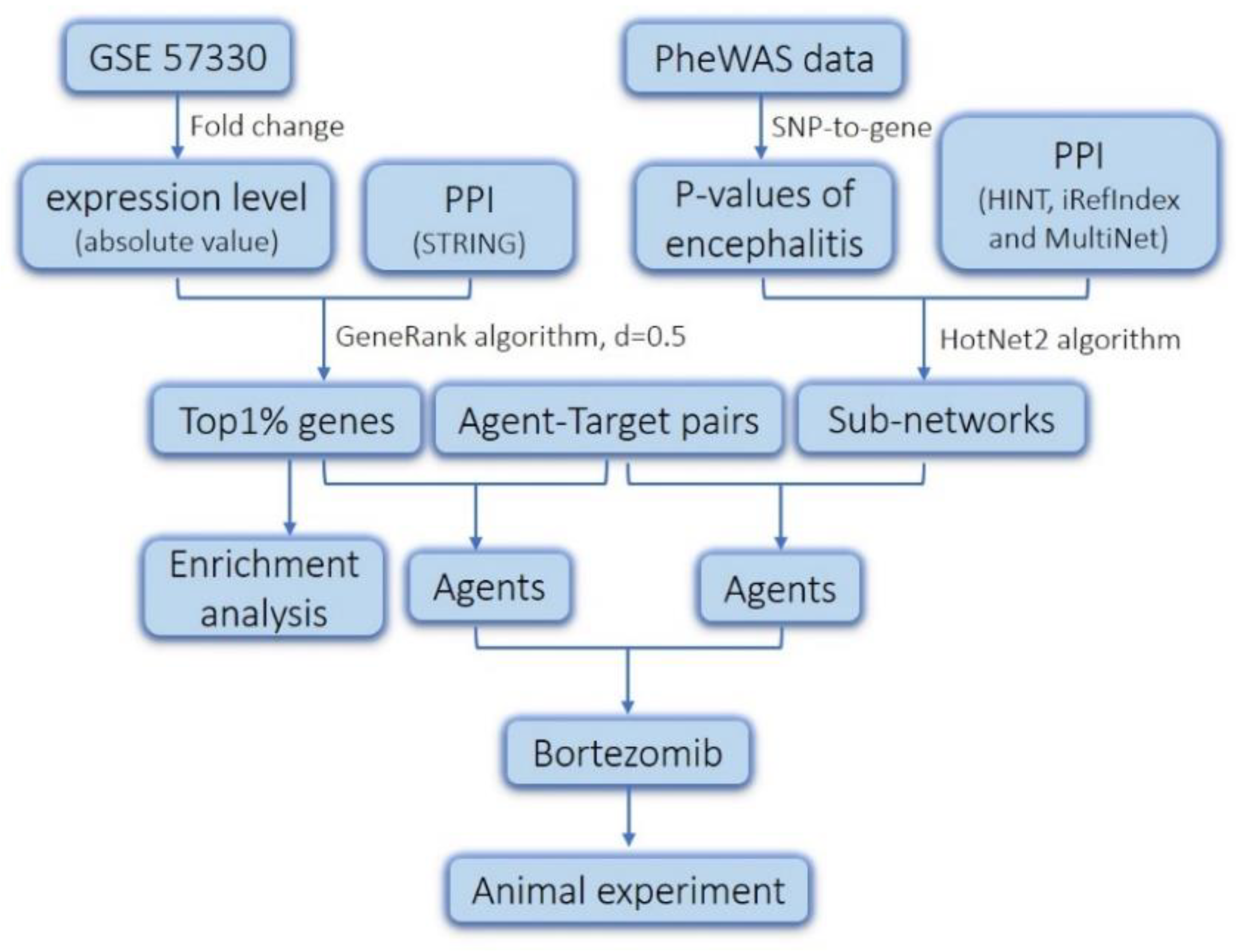
2.1. Screening of Genes Associated with JEV Infection by GeneRank Algorithm
2.2. Drug Repurposing for JEV Infection by Targeting GeneRank-Derived Genes
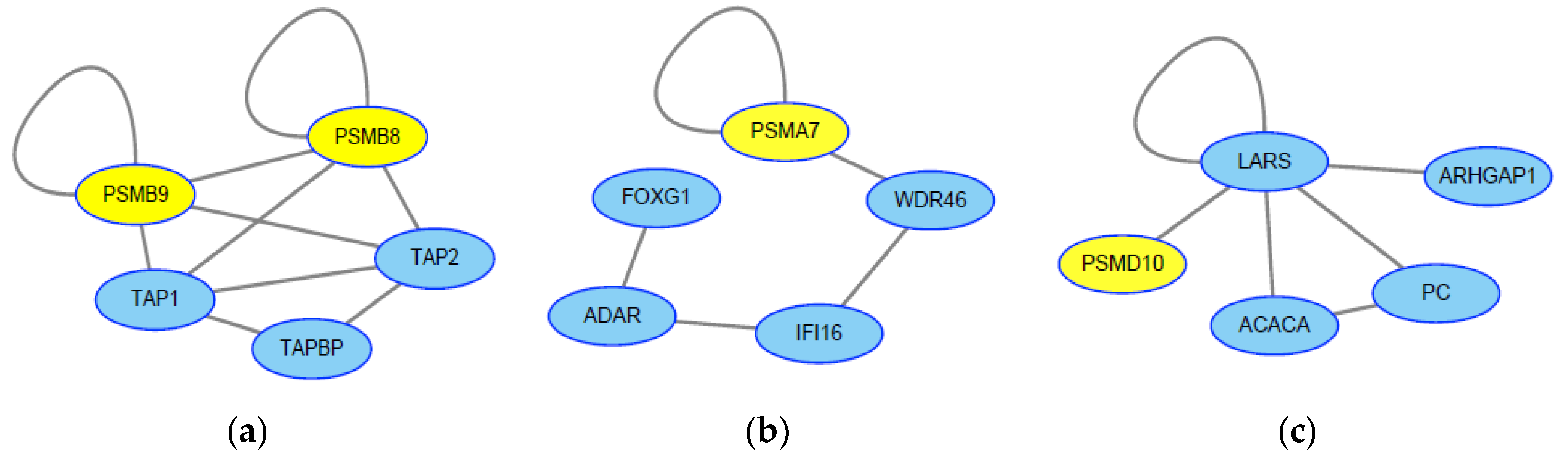
2.3. Screening of Genes Associated with JEV Infection by the HotNet2 Algorithm
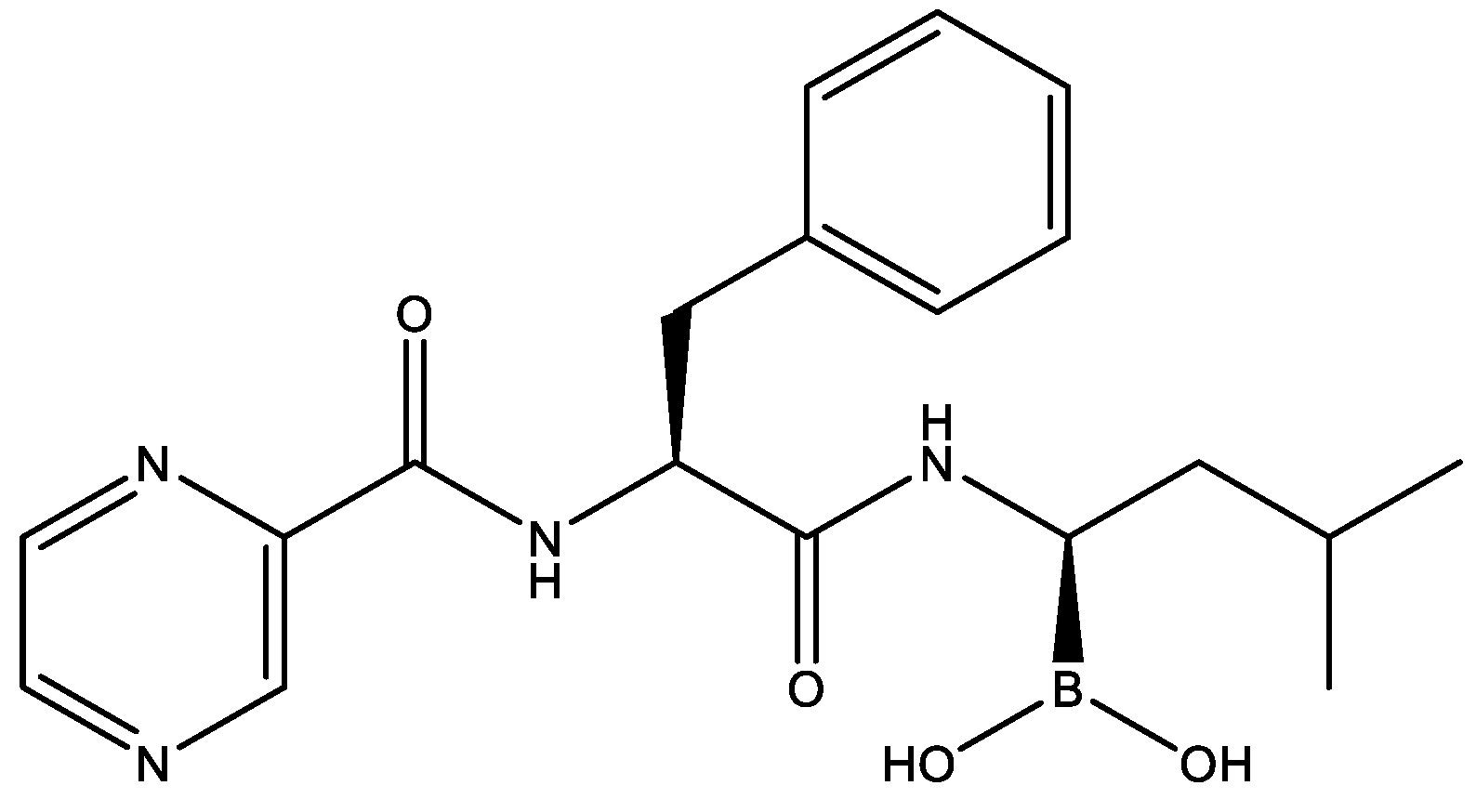
2.4. Drug Repurposing for JEV Infection by Targeting HotNet2-Derived Genes
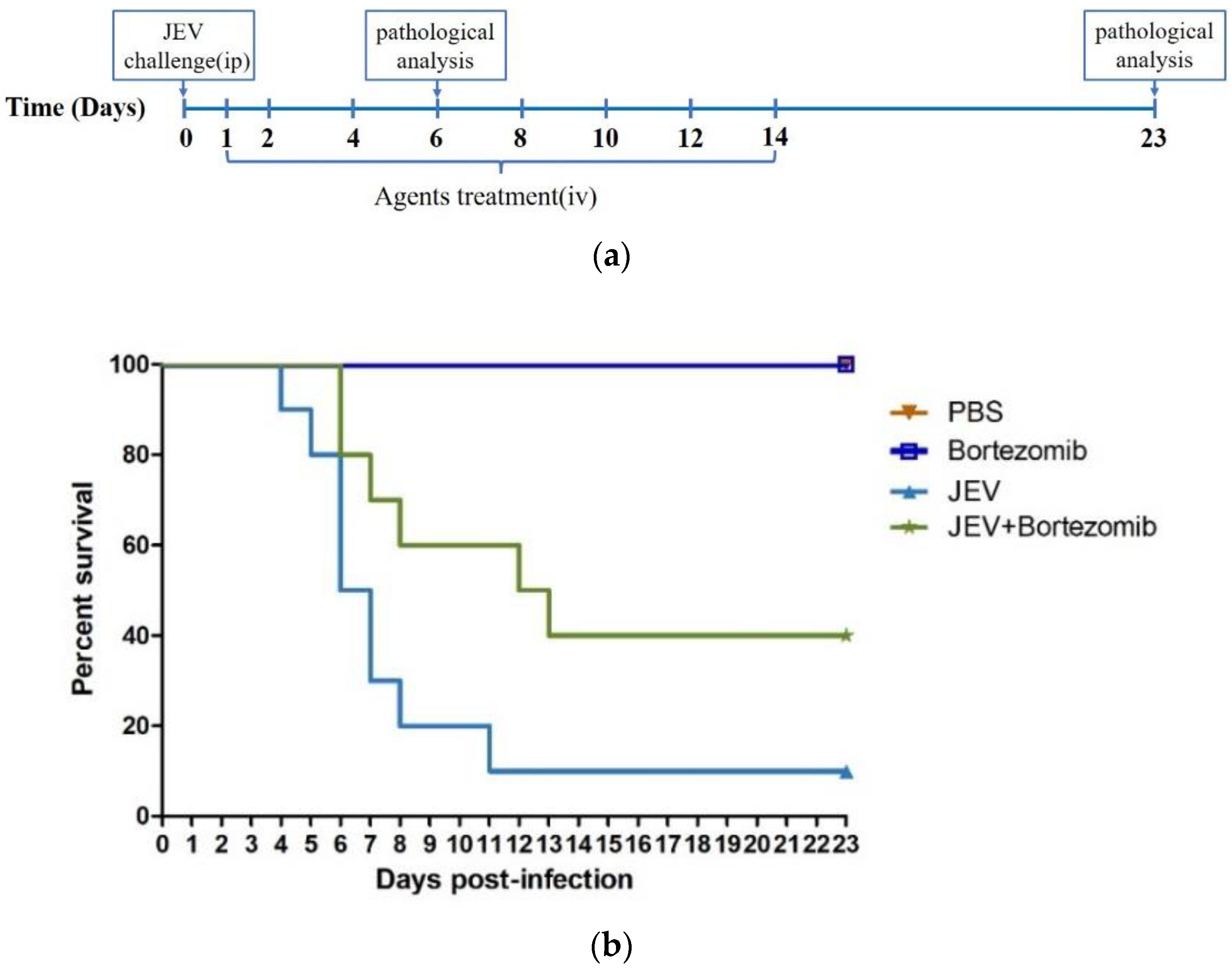
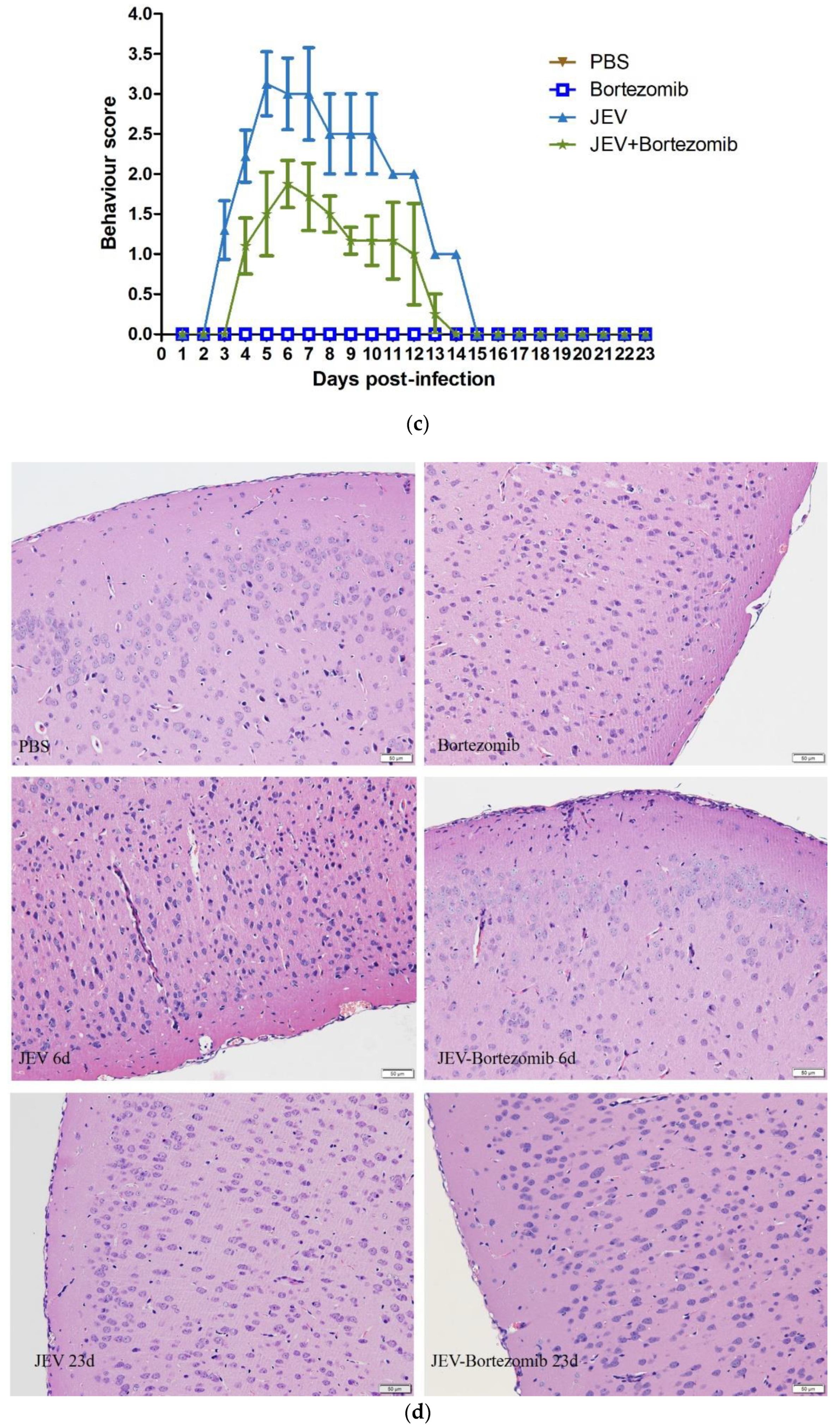
2.5. Therapeutic Effects of Bortezomib on JEV-Infected Mice
3. Conclusions
4. Materials and Methods
4.1. Data Resources
4.2. GeneRank Algorithm
4.3. HotNet2 Algorithm
4.4. Agents and Virus
4.5. Animal Studies
4.6. H&E Staining
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DMSO | dimethyl sulfoxide |
| PBS | phosphate buffer saline |
| PEG300 | polyethylene glycol 300 |
| PFU | plaque forming unit |
References
- Turtle, L.; Solomon, T. Japanese encephalitis—The prospects for new treatments. Nat. Rev. Neurol. 2018, 14, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.I.; Lee, Y.M. Japanese encephalitis: The virus and vaccines. Hum. Vaccin. Immunother. 2014, 10, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Endy, T.P.; Nisalak, A. Japanese Encephalitis Virus: Ecology and Epidemiology. Curr. Top. Microbiol. Immunol. 2002, 267, 11–48. [Google Scholar] [PubMed]
- Campbell, G.L.; Hills, S.L.; Fischer, M.; Jacobson, J.A.; Hoke, C.H.; Hombach, J.M.; Marfin, A.A.; Solomon, T.; Tsai, T.F.; Tsu, V.D. Estimated global incidence of Japanese encephalitis: A systematic review. Bull. World Health Org. 2015, 89, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Paulke-Korinek, M.; Kollaritsch, H. Japanese encephalitis and vaccines: Past and future prospects. Wien. Wochenschr. 2008, 120, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.H.; Bunnage, M.E. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 2017, 16, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, S.M. Overcoming drug development bottlenecks with repurposing: Old drugs learn new tricks. Nat. Med. 2014, 20, 590–591. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Saez-Rodriguez, J.; Bernardo, D. Di network based elucidation of drug response: From modulators to targets. BMC Syst. Biol. 2013, 7, 139. [Google Scholar] [CrossRef] [PubMed]
- Mercorelli, B.; Palù, G.; Loregian, A. Drug Repurposing for Viral Infectious Diseases: How Far Are We? Trends Microbiol. 2018, 26, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Schuler, J.; Hudson, M.L.; Schwartz, D.; Samudrala, R. A Systematic Review of Computational Drug Discovery, Development, and Repurposing for Ebola Virus Disease Treatment. Molecules 2017, 22, 1777. [Google Scholar] [CrossRef] [PubMed]
- Leal, E.S.; Aucar, M.G.; Gebhard, L.G.; Iglesias, N.G.; Pascual, M.J.; Casal, J.J.; Gamarnik, A.V.; Cavasotto, C.N.; Bollini, M. Discovery of novel dengue virus entry inhibitors via a structure-based approach. Bioorg. Med. Chem. Lett. 2017, 27, 3851–3855. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Xiong, L.; Chen, J.; Zhang, H.Y. Genetics-directed drug discovery for combating Mycobacterium tuberculosis infection. J. Biomol. Struct. Dyn. 2017, 35, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, B.; Zhu, J. Functional genomics- and network-driven systems biology approaches for pharmacogenomics and toxicogenomics. Curr. Drug Metabol. 2012, 13, 952–967. [Google Scholar] [CrossRef]
- Mei, H.; Xia, T.; Feng, G.; Zhu, J.; Lin, S.M.; Qiu, Y. Opportunities in systems biology to discover mechanisms and repurpose drugs for CNS diseases. Drug Discov. Today 2012, 17, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.L.; Breitling, R.; Higham, D.J.; Gilbert, D.R. GeneRank: Using search engine technology for the analysis of microarray experiments. BMC Bioinform. 2005, 21, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Leiserson, M.D.M.; Vandin, F.; Wu, H.-T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2014, 47, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Kumari, B.; Jain, P.; Das, S.; Ghosal, S.; Hazra, B.; Trivedi, A.C.; Basu, A.; Chakrabarti, J.; Vrati, S.; Banerjee, A. Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese encephalitis virus in microglial cells. Sci. Rep. 2016, 6, 20263. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zhu, D.; Lian, X.; Liu, W.; Cao, R.; Chen, P. Porcine 2′,5′-oligoadenylate synthetases inhibit Japanese encephalitis virus replication in vitro. J. Med. Virol. 2015, 88, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Manocha, G.; Mishra, R.; Sharma, N.; Kumawat, K.; Basu, A.; Singh, S.K. Regulatory role of TRIM21 in the type-I interferon pathway in Japanese encephalitis virus-infected human microglial cells. J. Neuroinflamm. 2014, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Wu, M.; Qian, S.; Zhou, Y.; Chen, H.; Li, X.; Qian, P. TRIM52 inhibits Japanese Encephalitis Virus replication by degrading the viral NS2A. Sci. Rep. 2016, 6, 33698–33709. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. Coruña J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.H.; Coffman, A.C.; Ainscough, B.J.; Spies, N.C.; Skidmore, Z.L.; Campbell, K.M.; Kilannin, K.; Pan, D.; Mcmichael, J.F.; Eldred, J.M. DGIdb 2.0: Mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016, 44, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhang, C.; Zhu, F.; Xu, F.; Chen, S.Y.; Zhang, P.; Li, Y.H.; Yang, S.Y.; Wei, Y.Q.; Tao, L. Therapeutic target database update 2014: A resource for targeted therapeutics. Nucleic Acids Res. 2014, 42, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Choy, M.M.; Zhang, S.L.; Costa, V.V.; Tan, H.C.; Horrevorts, S.; Ooi, E.E. Proteasome inhibition suppresses dengue virus egress in antibody dependent infection. PLOS Negl. Trop. Dis. 2015, 9, e0004058. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.L.; Deng, C.L.; Chen, X.; Wang, J.; Wang, S.B.; Wang, W.; Deng, F.; Zhang, B.; Xiao, G.; Zhang, L.K. Quantitative proteomic analysis of mosquito C6/36 cells reveals host proteins involved in Zika virus infection. J. Virol. 2017, 91, 00554–00571. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Raung, S.L.; Kuo, M.D.; Wang, Y.M. Suppression of Japanese encephalitis virus infection by non-steroidal anti-inflammatory drugs. J. Gen. Virol. 2002, 83, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Nawa, M. Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J. Gen. Virol. 2003, 84, 1737–1741. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Ghosh, D.; Basu, A. Curcumin protects neuronal cells from Japanese encephalitis virus-mediated cell death and also inhibits infective viral particle formation by dysregulation of ubiquitin–proteasome system. J. Neuroimmune Pharmacol. 2009, 4, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Jiang, R.; Cui, M.; Zhu, B.; Sun, L.; Wang, Y.; Zohaib, A.; Dong, Q.; Ruan, X.; Song, Y.; et al. Etanercept reduces neuroinflammation and lethality in mouse model of Japanese encephalitis. J. Infect. Dis. 2014, 210, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Raung, S.-L.; Chen, S.-Y.; Liao, S.-L.; Chen, J.-H.; Chen, C.-J. Tyrosine kinase inhibitors attenuate Japanese encephalitis virus-induced neurotoxicity. Biochem. Biophys. Res. Commun. 2005, 327, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.K.; Basu, A. Minocycline neuroprotects, reduces microglial activation, inhibits caspase 3 induction, and viral replication following Japanese encephalitis. J. Neurochem. 2008, 105, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Johari, J.; Kianmehr, A.; Mustafa, M.R.; Abubakar, S.; Zandi, K. Antiviral activity of baicalein and quercetin against the Japanese encephalitis virus. Int. J. Mol. Sci. 2012, 13, 16785–16795. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, L.; Desai, A.; Yogeeswari, P.; Sriram, D.; Madhusudana, S.N.; Ravi, V. Combination of N-methylisatin-β-thiosemicarbazone derivative (SCH16) with ribavirin and mycophenolic acid potentiates the antiviral activity of SCH16 against Japanese encephalitis virus in vitro. Lett. Appl. Microbiol. 2012, 55, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.Y.; Chang, Y.C.; Hua, C.H.; Chuang, C.; Huang, S.H.; Kung, S.H.; Hour, M.J.; Lin, C.W. Tubacin, an HDAC6 Selective Inhibitor, Reduces the Replication of the Japanese Encephalitis Virus via the Decrease of Viral RNA Synthesis. Int. J. Mol. Sci. 2017, 18, 954. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Ou, Y.C.; Chang, C.Y.; Pan, H.C.; Liao, S.L.; Chen, S.Y.; Raung, S.L.; Lai, C.Y. Glutamate released by Japanese encephalitis virus-infected microglia involves TNF-α signaling and contributes to neuronal death. Glia 2012, 60, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tripathi, P.; Baranwal, M.; Singh, S.; Tripathi, S.; Banerjee, G. Randomized, controlled trial of oral ribavirin for Japanese encephalitis in children in Uttar Pradesh, India. Clin. Infect. Dis. 2009, 48, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Basu, A.; Sinha, S.; Das, M.; Tripathi, P.; Jain, A.; Kumar, C.; Atam, V.; Khan, S.; Singh, A.S. Role of oral Minocycline in acute encephalitis syndrome in India-a randomized controlled trial. BMC Infect. Dis. 2015, 16, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Tipney, H.; Painter, J.L.; Shen, J.; Nicoletti, P.; Shen, Y.; Floratos, A.; Sham, P.C.; Li, M.J.; Wang, J.; et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015, 47, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Bastarache, L.; Ritchie, M.D.; Carroll, R.J.; Zink, R.; Mosley, J.D.; Field, J.R.; Pulley, J.M.; Ramirez, A.H.; Bowton, E. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013, 31, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, H.; Zu, X.; Liu, Y.; Chen, L.; Zhu, X.; Zhang, L.; Zhou, Z.; Xiao, G.; Wang, W. The ubiquitin-proteasome system is essential for the productive entry of Japanese encephalitis virus. Virology 2016, 498, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.-D.; Meertens, L.; Bonazzi, M.; Cossart, P.; Arenzana-Seisdedos, F.; Amara, A. Appraising the roles of cbll1 and the ubiquitin/proteasome system for flavivirus entry and replication. J. Virol. 2010, 85, 2980–2989. [Google Scholar] [CrossRef] [PubMed]
- Amaya, M.; Keck, F.; Lindquist, M.; Voss, K.; Scavone, L.; Kehn-Hall, K.; Roberts, B.; Bailey, C.; Schmaljohn, C.; Narayanan, A. The ubiquitin proteasome system plays a role in venezuelan equine encephalitis virus infection. PLoS ONE 2015, 10, e0124792. [Google Scholar] [CrossRef] [PubMed]
- Starkey, V.G.; Kirk, C.A.V.; Bixler, G.V.; Imperio, C.G.; Kale, V.P.; Serfass, J.M.; Farley, J.A.; Yan, H.; Warrington, J.P.; Han, S. Neuroglial Expression of the MHCI Pathway and PirB Receptor Is Upregulated in the Hippocampus with Advanced Aging. J. Mol. Neurosci. 2012, 48, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.R.; Puig, M.; Darnell, M.E.R.; Mihalik, K.; Feinstone, S.M. New antiviral pathway that mediates hepatitis c virus replicon interferon sensitivity through adar1. J. Virol. 2005, 79, 6291–6298. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Liu, Z.; Kang, W.; Li, H.; Zhang, L.; Zhang, N. Interferon beta 1a versus interferon beta 1a plus ribavirin for the treatment of chronic hepatitis c in Chinese patients: A randomized, placebo-controlled trial. Dig. Dis. Sci. 2007, 53, 2238–2245. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yao, Y.; Yeung, M.-L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Batista, M.N.; Carneiro, B.M.; Braga, A.C.S.; Rahal, P. Caffeine inhibits hepatitis c virus replication in vitro. Arch. Virol. 2014, 160, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.A.; Feld, J.J.; Park, Y.; Kleiner, D.E.; Everhart, J.E.; Liang, T.J.; Hoofnagle, J.H. Increased caffeine consumption is associated with reduced hepatic fibrosis. Hepatology 2009, 51, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Luthra, P.; Aguirre, S.; Yen, B.C.; Pietzsch, C.A.; Sanchez-Aparicio, M.T.; Tigabu, B.; Morlock, L.K.; García-Sastre, A.; Leung, D.W.; Williams, N.S. Topoisomerase II Inhibitors Induce DNA Damage-Dependent Interferon Responses Circumventing Ebola Virus Immune Evasion. mBio 2017, 8, e00368-17. [Google Scholar] [CrossRef] [PubMed]
- Deejai, N.; Roshorm, Y.M.; Kubera, A. Antiviral compounds against nucleocapsid protein of porcine epidemic diarrhea virus. Anim. Biotechnol. 2016, 28, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.B.; Klaus, J.O.; Keith, S.G. Carfilzomib: A second-generation proteasome inhibitor for the treatment of multiple myeloma. Am. Soc. Health Syst. Pharm. 2015, 72, 353–360. [Google Scholar]
- Scheinfeld, N. A review of deferasirox, bortezomib, dasatinib, and cyclosporine eye drops: Possible uses and known side effects in cutaneous medicine. J. Drugs Dermatol. 2007, 6, 352–355. [Google Scholar] [PubMed]
- Das, J.; Yu, H. Hint: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 2012, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Razick, S.; Magklaras, G.; Donaldson, I.M. Irefindex: A consolidated protein interaction database with provenance. BMC Bioinform. 2008, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Fu, Y.; Chen, J.; Gerstein, M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput. Biol. 2013, 9, e1002886. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; Kuhn, M.; Bork, P.; Jensen, L.J.; von Mering, C. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Agent | Anti-JEV Potential | Reference |
|---|---|---|
| Aspirin | Aspirin suppressed JEV propagation in neuronal and nonneuronal cells | [27] |
| Chlorpromazine | Chlorpromazine reduced the positive rate of JEV infection by 50% in vitro | [28] |
| Curcumin | Curcumin inhibited the production of infective JEV particle in vitro | [29] |
| Etanercept | Etanercept significantly relieved clinical symptoms and reduces mortality in JEV-infected mice | [30] |
| Genistein | Genistein protected neurons from JEV-induced decrease in the number of visible neurons | [31] |
| Minocycline | Minocycline protected 70% of mice from JEV-induced death, and inhibited JEV replication in vitro | [32] |
| Quercetin | Quercetin inhibited JEV replication in vitro | [33] |
| Ribavirin | Ribavirin inhibited JEV replication in vitro | [34] |
| Valproic acid | Valproic acid reduced the cytopathic effects caused by JEV | [35] |
| Serial Number | Agents | Indications | Evidence in Antiviral |
|---|---|---|---|
| 1 | Amoxicillin | bacterial infections | N |
| 2 | AT-406 | cancer | N |
| 3 | Biotin | dietary shortage or imbalance | Y |
| 4 | Bortezomib | multiple myeloma, lymphoma | Y |
| 5 | Caffeine | fatigue, neurasthenia | Y |
| 6 | Carfilzomib | multiple myeloma | N |
| 7 | Clavulanate | bacterial infections | N |
| 8 | Doxorubicin | various cancer | Y |
| 9 | GDC-0152 | cancer | N |
| 10 | Glatiramer Acetate | multiple sclerosis | N |
| 11 | Insulin | diabetes | N |
| 12 | Interferon Beta-1A | multiple sclerosis, condyloma acuminatum | Y |
| 13 | Interferon Beta-1B | multiple sclerosis | Y |
| 14 | N-Acetylglucosamine | osteoarthritis | N |
| 15 | Niraparib | ovarian cancer, fallopian tube cancer, breast cancer | N |
| 16 | Olaparib | ovarian cancer, breast cancer | N |
| 17 | Pyruvic Acid | dietary shortage or imbalance | N |
| 18 | Rucaparib | ovarian cancer | N |
| 19 | Talazoparib | breast cancer | N |
| 20 | Veliparib | breast cancer, non-small cell lung cancer | N |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, B.-M.; Tong, X.-Y.; Quan, Y.; Liu, M.-Y.; Zhang, Q.-Y.; Song, Y.-F.; Zhang, H.-Y. Drug Repurposing for Japanese Encephalitis Virus Infection by Systems Biology Methods. Molecules 2018, 23, 3346. https://doi.org/10.3390/molecules23123346
Lv B-M, Tong X-Y, Quan Y, Liu M-Y, Zhang Q-Y, Song Y-F, Zhang H-Y. Drug Repurposing for Japanese Encephalitis Virus Infection by Systems Biology Methods. Molecules. 2018; 23(12):3346. https://doi.org/10.3390/molecules23123346
Chicago/Turabian StyleLv, Bo-Min, Xin-Yu Tong, Yuan Quan, Meng-Yuan Liu, Qing-Ye Zhang, Yun-Feng Song, and Hong-Yu Zhang. 2018. "Drug Repurposing for Japanese Encephalitis Virus Infection by Systems Biology Methods" Molecules 23, no. 12: 3346. https://doi.org/10.3390/molecules23123346
APA StyleLv, B.-M., Tong, X.-Y., Quan, Y., Liu, M.-Y., Zhang, Q.-Y., Song, Y.-F., & Zhang, H.-Y. (2018). Drug Repurposing for Japanese Encephalitis Virus Infection by Systems Biology Methods. Molecules, 23(12), 3346. https://doi.org/10.3390/molecules23123346





