Effects of Limited Hydrolysis and High-Pressure Homogenization on Functional Properties of Oyster Protein Isolates
Abstract
:1. Introduction
2. Results and Discussion
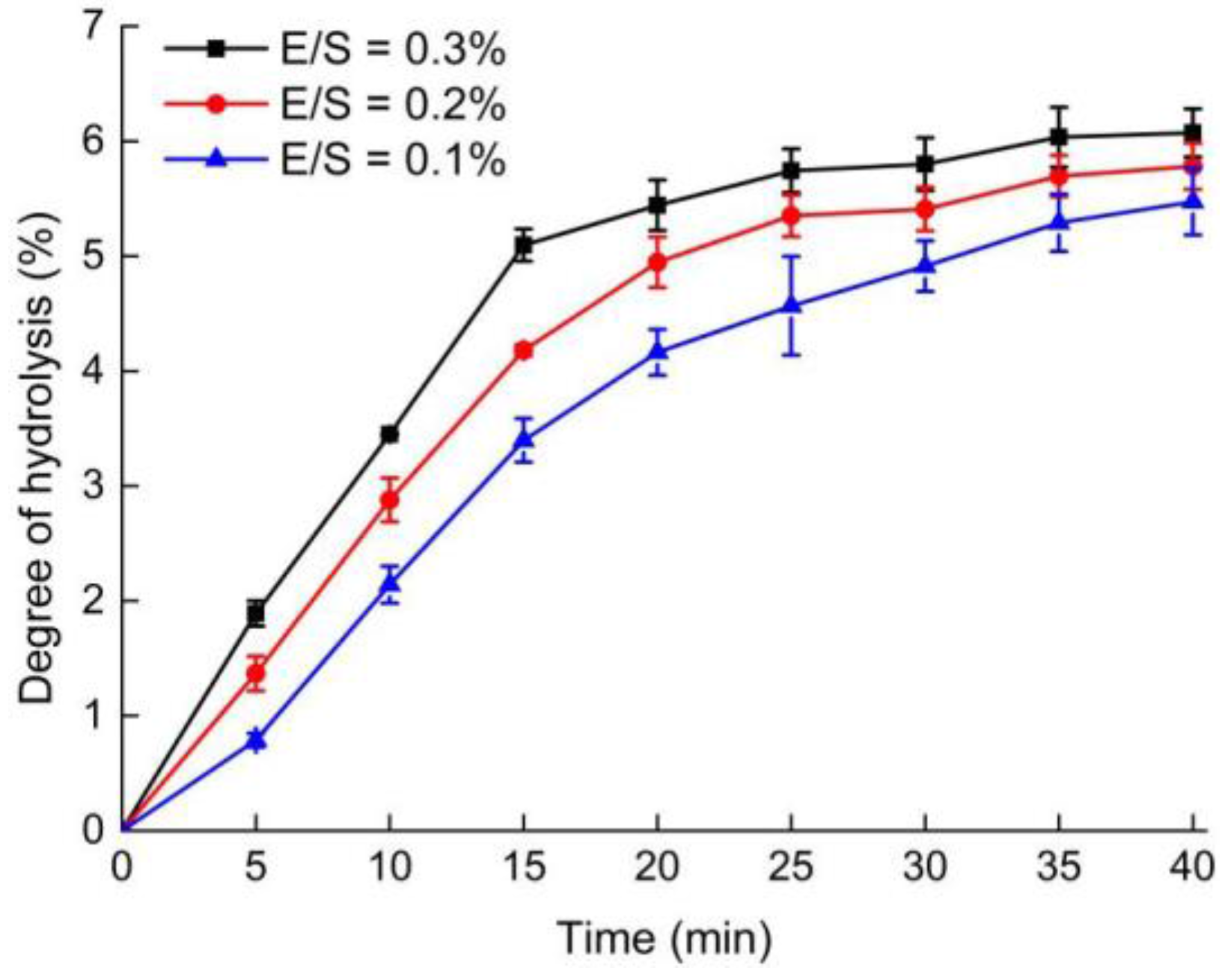
2.1. Degree of Hydrolysis (DH) of OPI Hydrolysis with Different Ratios of Enzyme to Substrate (E/S)
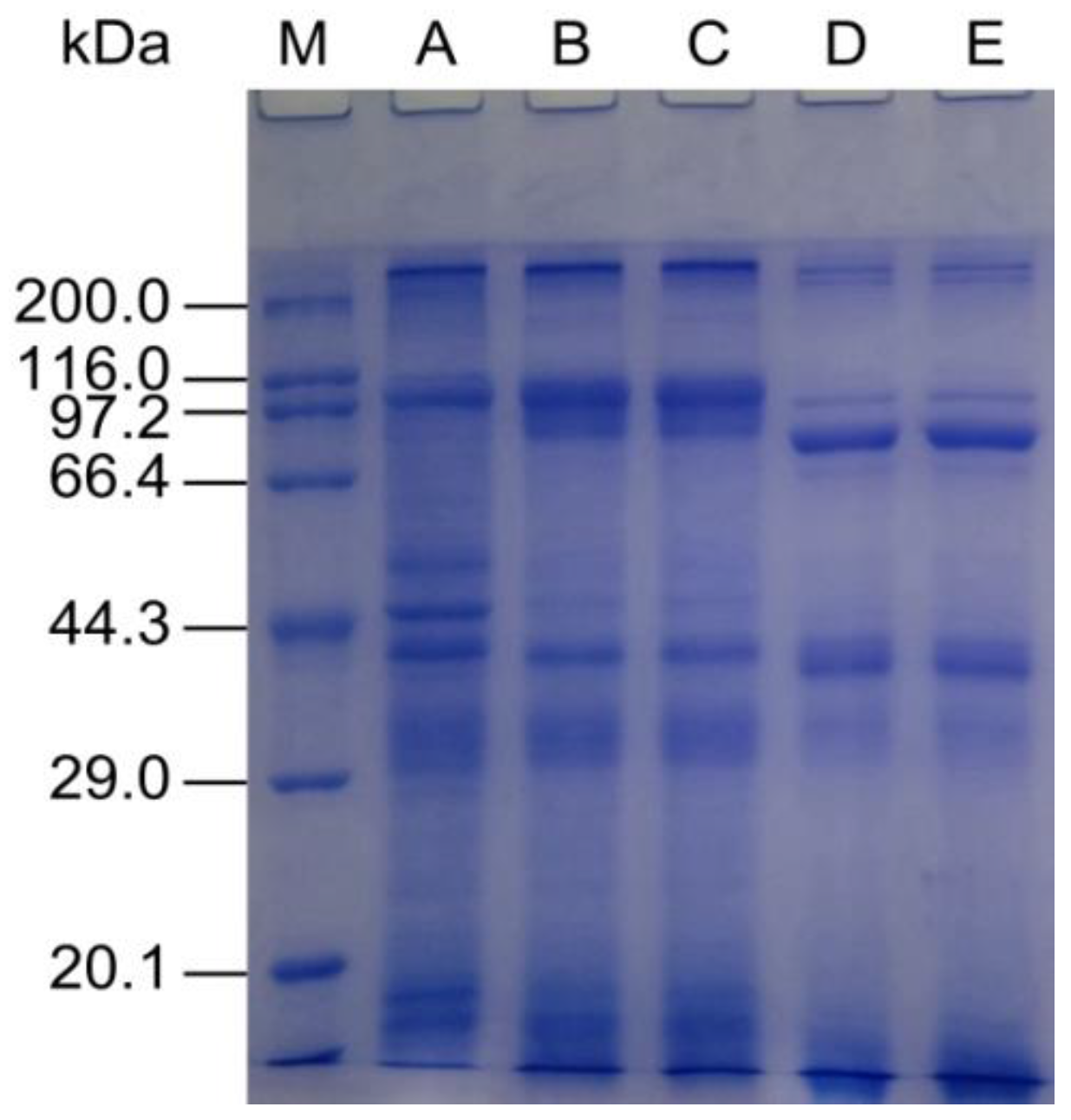
2.2. Composition and SDS-PAGE Analysis of Protein Isolates and Hydrolysates
2.3. Protein Solubility
2.4. Particle Size Distribution and Zeta Potential
2.5. Surface Hydrophobicity
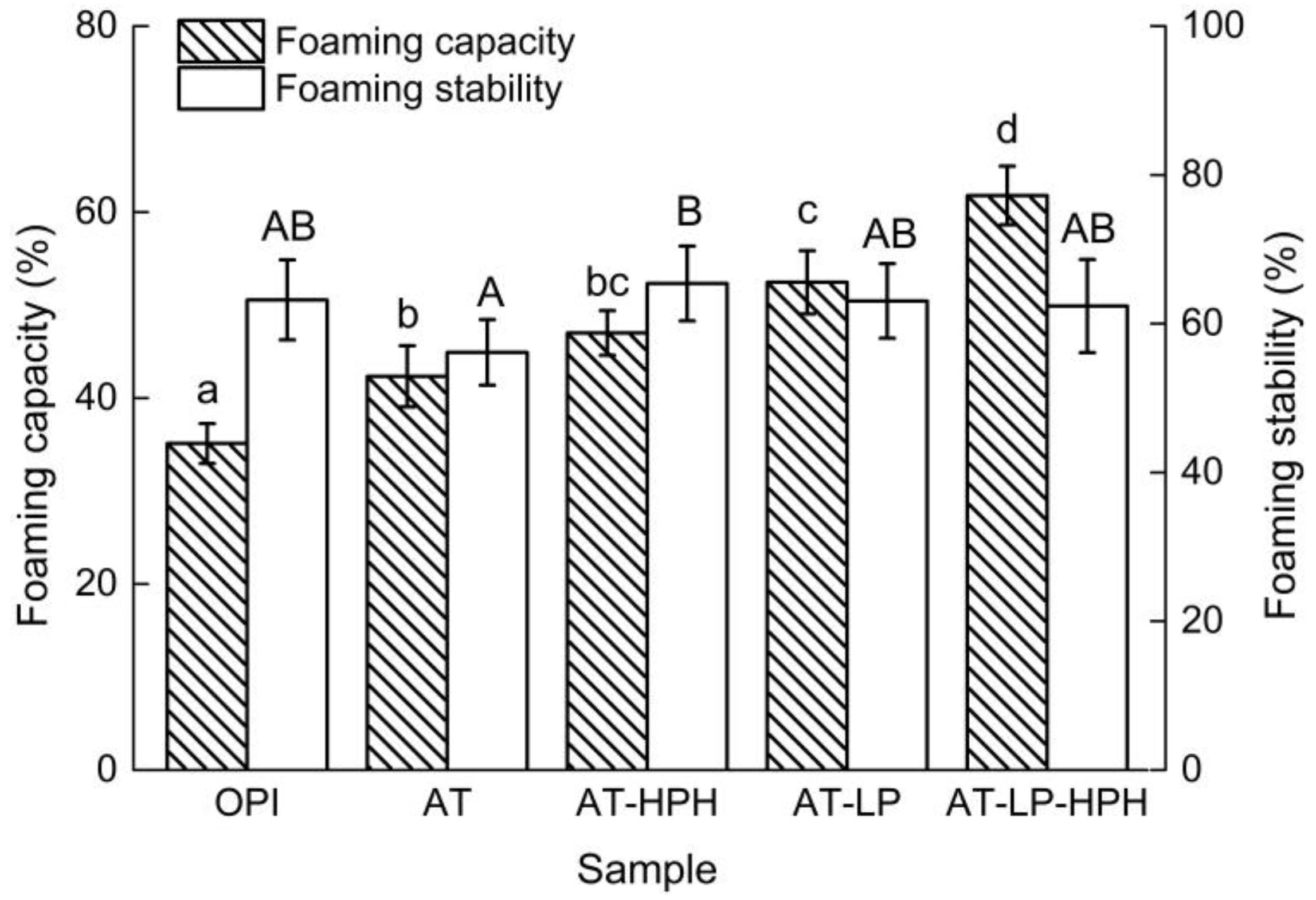
2.6. Foaming Properties
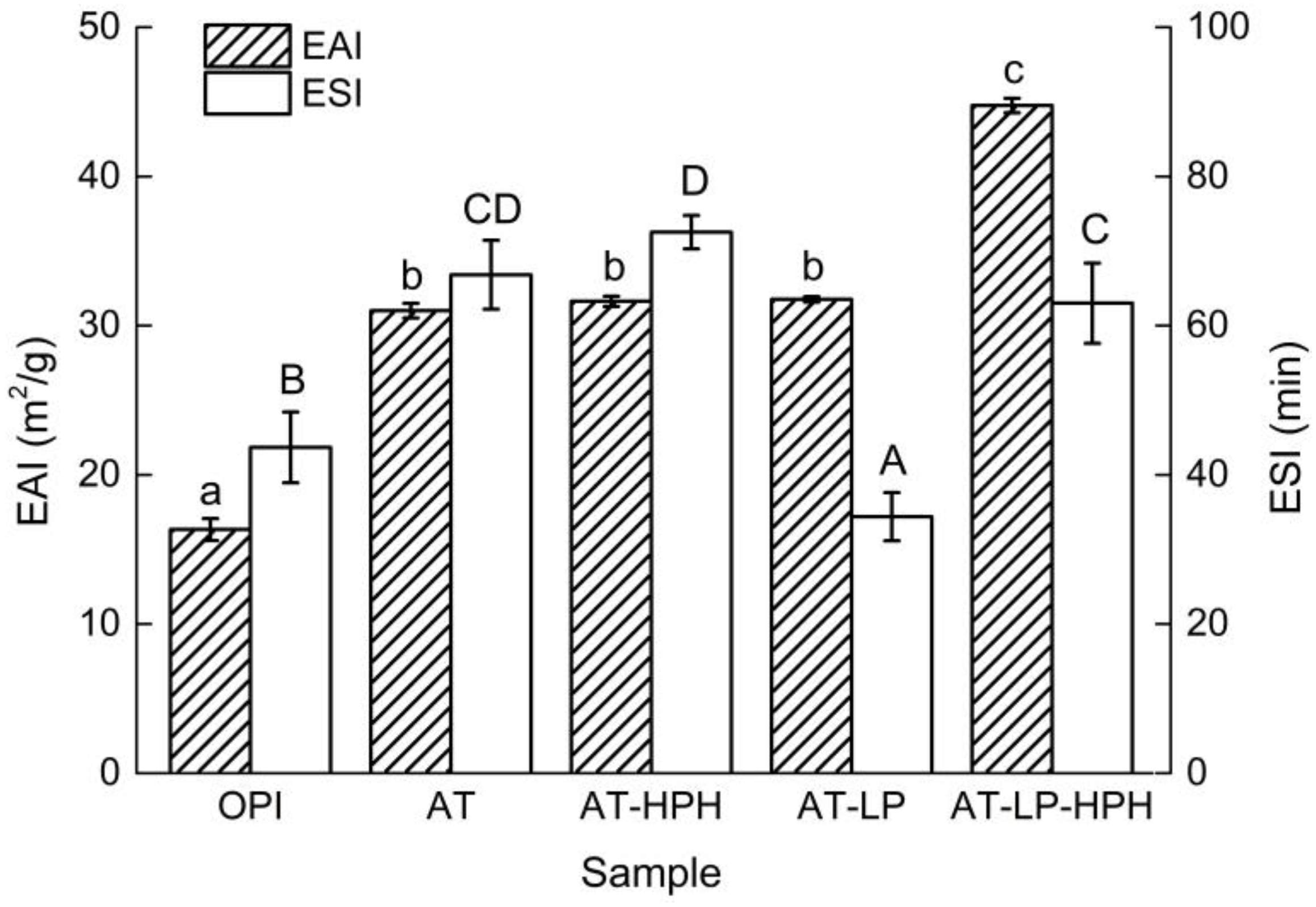
2.7. Emulsifying Properties
3. Materials and Methods
3.1. Chemical Reagents
3.2. Determination of Composition of OPI
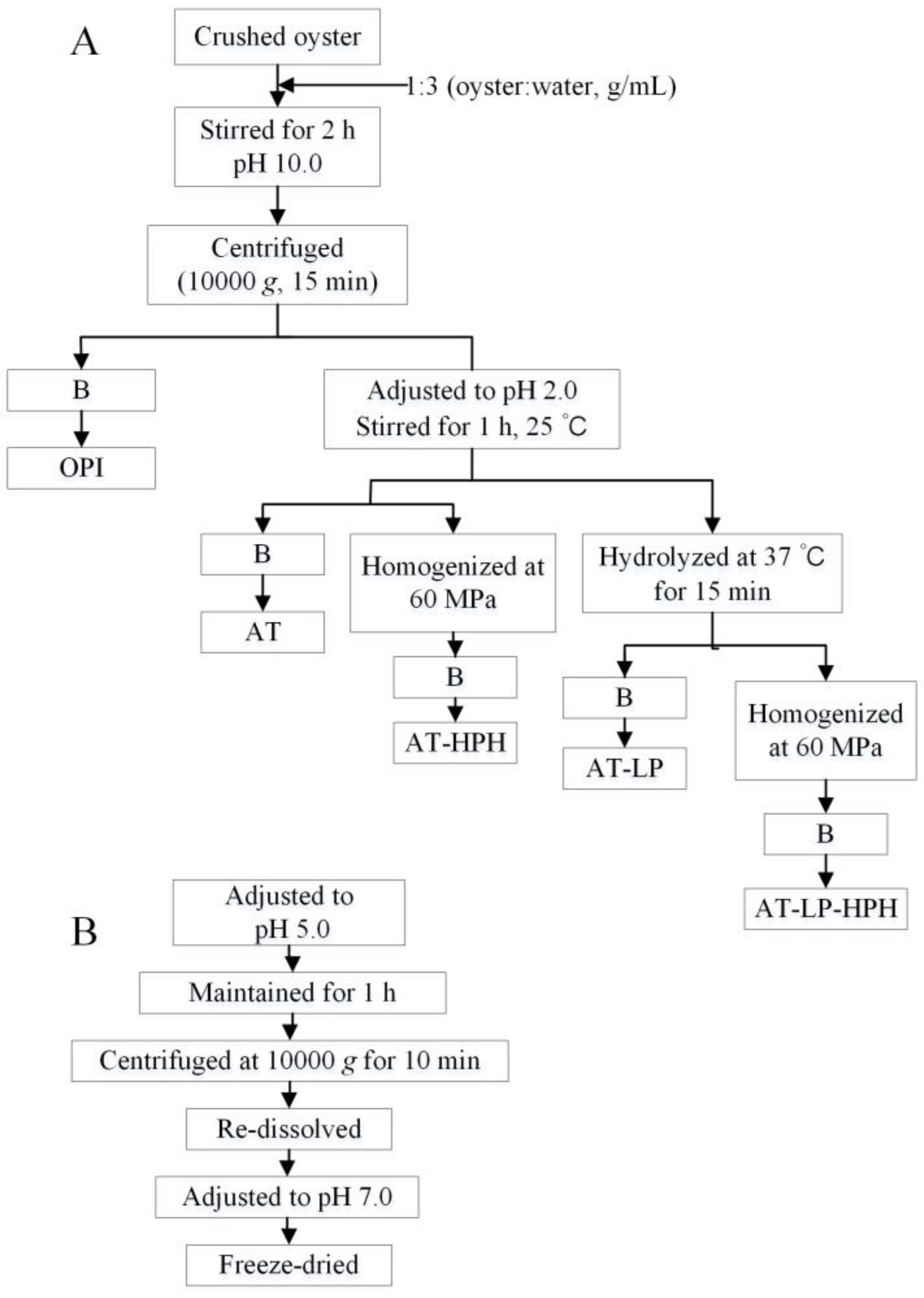
3.3. Preparation of Protein Isolates and Hydrolysates
3.4. Determination of Degree of Hydrolysis (DH)
3.5. SDS-PAGE Analysis
3.6. Determination of Protein Solubility
3.7. Determination of Particle Size Distribution And Zeta Potential
3.8. Determination of Surface Hydrophobicity (H0)
3.9. Determination of Foaming Properties
3.10. Determination of Emulsifying Properties
3.11. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hayes, M.; Skomedal, H.; Skjånes, K.; Mazur-Marzec, H.; Toruńska-Sitarz, A.; Catala, M.; Isleten Hosoglu, M.; García-Vaquero, M. 15—Microalgal proteins for feed, food and health A2—Gonzalez-Fernandez, Cristina. In Microalgae-Based Biofuels and Bioproducts; Muñoz, R., Ed.; Woodhead Publishing: Cambridge, UK, 2017; pp. 347–368. [Google Scholar]
- Ventura, S.P.M.; Nobre, B.P.; Ertekin, F.; Hayes, M.; Garciá-Vaquero, M.; Vieira, F.; Koc, M.; Gouveia, L.; Aires-Barros, M.R.; Palavra, A.M.F. 19—Extraction of value-added compounds from microalgae A2—Gonzalez-Fernandez, Cristina. In Microalgae-Based Biofuels and Bioproducts; Muñoz, R., Ed.; Woodhead Publishing: Cambridge, UK, 2017; pp. 461–483. [Google Scholar]
- Miranda, M.; Lopez-Alonso, M.; Garcia-Vaquero, M. Macroalgae for Functional Feed Development: Applications in Aquaculture, Ruminant and Swine Feed Industries; NOVA Science Publishers: Hauppauge, NY, USA, 2017. [Google Scholar]
- Barba, F.J. Microalgae and seaweeds for food applications: Challenges and perspectives. Food Res. Int. 2017, 99, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Rajauria, G.; Jaiswal, A.K.; Abu-Gannam, N.; Gupta, S. Antimicrobial, antioxidant and free radical-scavenging capacity of brown seaweed himanthalia elongata from western coast of ireland. J. Food Biochem. 2013, 37, 322–335. [Google Scholar] [CrossRef]
- Grienke, U.; Silke, J.; Tasdemir, D. Bioactive compounds from marine mussels and their effects on human health. Food Chem. 2014, 142, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Harada, R. Proximate and amino acid composition of the roe and muscle of selected marine species. J. Food Sci. 1985, 50, 1585–1587. [Google Scholar] [CrossRef]
- Yu, C.; Cha, Y.; Wu, F.; Xu, X.; Qin, Y.; Li, X.; Du, M. Effects of high-pressure homogenisation on structural and functional properties of mussel (Mytilus edulis) protein isolate. Int. J. Food Sci. Technol. 2017. [Google Scholar] [CrossRef]
- Galazka, V.B.; Dickinson, E.; Ledward, D.A. Influence of high pressure processing on protein solutions and emulsions. Curr. Opin. Colloid Interface Sci. 2000, 5, 182–187. [Google Scholar] [CrossRef]
- Balny, C.; Masson, P. Effects of high pressure on proteins. Food Rev. Int. 1993, 9, 611–628. [Google Scholar] [CrossRef]
- Yuan, B.; Ren, J.; Zhao, M.; Luo, D.; Gu, L. Effects of limited enzymatic hydrolysis with pepsin and high-pressure homogenization on the functional properties of soybean protein isolate. LWT Food Sci. Technol. 2012, 46, 453–459. [Google Scholar] [CrossRef]
- Luo, D.; Zhao, Q.; Zhao, M.; Yang, B.; Long, X.; Ren, J.; Zhao, H. Effects of limited proteolysis and high-pressure homogenisation on structural and functional characteristics of glycinin. Food Chem. 2010, 122, 25–30. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.; Ren, J.; Zhao, M. Effects of ultrasound pretreatment on the enzymatic hydrolysis of soy protein isolates and on the emulsifying properties of hydrolysates. J. Agric. Food Chem. 2011, 59, 2600–2609. [Google Scholar] [CrossRef] [PubMed]
- Peñas, E.; Préstamo, G.; Gomez, R. High pressure and the enzymatic hydrolysis of soybean whey proteins. Food Chem. 2004, 85, 641–648. [Google Scholar] [CrossRef]
- Panyam, D.; Kilara, A. Enhancing the functionality of food proteins by enzymatic modification. Trends Food Sci. Technol. 1996, 7, 120–125. [Google Scholar] [CrossRef]
- Jung, S.; Murphy, P.A.; Johnson, L.A. Physicochemical and functional properties of soy protein substrates modified by low levels of protease hydrolysis. J. Food Sci. 2005, 70, C180–C187. [Google Scholar] [CrossRef]
- Ventureira, J.L.; Martínez, E.N.; Añón, M.C. Effect of acid treatment on structural and foaming properties of soy amaranth protein mixtures. Food Hydrocoll. 2012, 29, 272–279. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Lopez-Alonso, M.; Hayes, M. Assessment of the functional properties of protein extracted from the brown seaweed Himanthalia elongata (Linnaeus) S. F. Gray. Food Res. Int. 2017, 99, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, S.L.; Gauthier, S.F.; Paquin, P. Emulsifying property of whey peptide fractions as a function of ph and ionic strength. J. Food Sci. 1992, 57, 601–604. [Google Scholar] [CrossRef]
- Tsumura, K.; Saito, T.; Tsuge, K.; Ashida, H.; Kugimiya, W.; Inouye, K. Functional properties of soy protein hydrolysates obtained by selective proteolysis. LWT Food Sci. Technol. 2005, 38, 255–261. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, R.; Xu, X.; Zhou, G.; Liu, D. Structural modification by high-pressure homogenization for improved functional properties of freeze-dried myofibrillar proteins powder. Food Res. Int. 2017, 100, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chen, J.; Xiong, Y.L. Structural and emulsifying properties of soy protein isolate subjected to acid and alkaline pH-shifting processes. J. Agric. Food Chem. 2009, 57, 7576–7583. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, K.; Srinivas, H. Controlled enzymatic hydrolysis of glycinin: Susceptibility of acidic and basic subunits to proteolytic enzymes. LWT Food Sci. Technol. 2007, 40, 1056–1065. [Google Scholar] [CrossRef]
- Song, X.; Zhou, C.; Fu, F.; Chen, Z.; Wu, Q. Effect of high-pressure homogenization on particle size and film properties of soy protein isolate. Ind. Crops Prod. 2013, 43, 538–544. [Google Scholar] [CrossRef]
- Fievet, P.; Szymczyk, A.; Labbez, C.; Aoubiza, B.; Simon, C.; Foissy, A.; Pagetti, J. Determining the zeta potential of porous membranes using electrolyte conductivity inside pores. J. Colloid Interface Sci. 2001, 235, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Adler-Nissen, J. In enzymatic hydrolysis of food proteins. Can. Med. Assoc. J. 1986, 172, 1783–1785. [Google Scholar]
- Chen, X.; Xu, X.; Zhou, G. Potential of high pressure homogenization to solubilize chicken breast myofibrillar proteins in water. Innov. Food Sci. Emerg. Technol. 2016, 33, 170–179. [Google Scholar] [CrossRef]
- Surówka, K.; Żmudziński, D.; Surówka, J. Enzymic modification of extruded soy protein concentrates as a method of obtaining new functional food components. Trends Food Sci. Technol. 2004, 15, 153–160. [Google Scholar] [CrossRef]
- Vioque, J.; Sánchez-Vioque, R.; Clemente, A.; Pedroche, J.; Millán, F. Partially hydrolyzed rapeseed protein isolates with improved functional properties. J. Am. Oil Chem. Soc. 2000, 77, 447–450. [Google Scholar] [CrossRef]
- Wang, M.; Hettiarachchy, N.S.; Qi, M.; Burks, W.; Siebenmorgen, T. Preparation and functional properties of rice bran protein isolate. J. Agric. Food Chem. 1999, 47, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, T. Egg yolk protein modification by controlled enzymatic hydrolysis for improved functionalities. Int. J. Food Sci. Technol. 2009, 44, 763–769. [Google Scholar] [CrossRef]
- Goff, H.D.; Kinsella, J.E.; Jordan, W.K. Influence of various milk protein isolates on ice cream emulsion stability. J. Dairy Sci. 1989, 72, 385–397. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Fish protein hydrolysates: Production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef] [PubMed]
- Haard, N.F. Enzymatic Modification of Protein in Food System; Technomic Publishing Co. Inc.: New York, NY, USA, 2001; pp. 155–190. [Google Scholar]
- Wagner, J.R.; Guéguen, J. Surface functional properties of native, acid-treated, and reduced soy glycinin. 2. emulsifying properties. J. Agric. Food Chem. 1999, 47, 2181–2187. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.M.; Mu, T.H.; Zhang, M.; Arogundade, L.A. The effects of pH and high hydrostatic pressure on the physicochemical properties of a sweet potato protein emulsion. Food Hydrocoll. 2014, 35, 209–216. [Google Scholar] [CrossRef]
- International, A. Official methods of analysis of AOAC International, 16th edition. Volume 1. Trends Food Sci. Technol. 1995, 6, 382–383. [Google Scholar]
- Carroll, N.V.; Longley, R.W.; Roe, J.H. The determination of glycogen in liver and muscle by use of anthrone reagent. J. Biol. Chem. 1956, 220, 583. [Google Scholar] [PubMed]
- Hooijerink, H.; Bennekom, E.O.V.; Nielen, M.W.F. Screening for gestagens in kidney fat using accelerated solvent extraction and liquid chromatography electrospray tandem mass spectrometry. Anal. Chim. Acta 2003, 483, 51–59. [Google Scholar] [CrossRef]
- Vorm, P.D.J.V.D. Dry ashing of plant material and dissolution of the ash in HF for the colorimetric determination of silicon. Commun. Soil Sci. Plant Anal. 1987, 18, 1181–1189. [Google Scholar] [CrossRef]
- Puppo, C.; Chapleau, N.; Speroni, F.; de Lamballerie-Anton, M.; Michel, F.; Añón, C.; Anton, M. Physicochemical modifications of high-pressure-treated soybean protein isolates. J. Agric. Food Chem. 2004, 52, 1564–1571. [Google Scholar] [CrossRef] [PubMed]
- Deak, N.A.; Murphy, P.A.; Johnson, L.A. Characterization of fractionated soy proteins produced by a new simplified procedure. J. Am. Oil Chem. Soc. 2006, 84, 137. [Google Scholar] [CrossRef]
- Spellman, D.; McEvoy, E.; O’Cuinn, G.; FitzGerald, R.J. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the TNBS, OPA and pH stat methods for quantification of degree of hydrolysis. Int. Dairy J. 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007, 102, 1317–1327. [Google Scholar] [CrossRef]
- Goa, J. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand. J. Clin. Lab. Investig. 1953, 5, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Song, Y.; Frank, P.; Evans, A.M. Particle size reduction to the nanometer range: A promising approach to improve buccal absorption of poorly water-soluble drugs. Int. J. Nanomed. 2011, 6, 1245–1251. [Google Scholar]
- Ju, Z.Y.; Hettiarachchy, N.S.; Rath, N. Extraction, denaturation and hydrophobic Properties of Rice Flour Proteins. J. Food Sci. 2001, 66, 229–232. [Google Scholar] [CrossRef]
- Liceaga-Gesualdo, A.M.; Li-Chan, E.C.Y. Functional properties of fish protein hydrolysate from herring (Clupea harengus). J. Food Sci. 2010, 64, 1000–1004. [Google Scholar] [CrossRef]
- Karaca, A.C.; Low, N.; Nickerson, M. Emulsifying properties of canola and flaxseed protein isolates produced by isoelectric precipitation and salt extraction. Food Res. Int. 2011, 44, 2991–2998. [Google Scholar] [CrossRef]
Sample Availability: Not available. |

| Samples | pH Levels | |||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| OPI | 54.4% ± 1.3% d | 3.9% ± 0.6% a | 2.6% ± 0.6% a | 7.6% ± 1.4% a | 27.1% ± 0.7% b | 27.4% ± 1.4% b | 41.5% ± 1.3% b | 59.8% ± 0.5% c |
| AT | 41.5% ± 0.8% b | 4.1% ± 0.7% a | 2.3% ± 0.5% a | 6.5% ± 1.3% a | 19.9% ± 0.8% a | 24.4% ± 1.5% b | 35.3% ± 1.1% a | 49.3% ± 0.6% a |
| AT-HPH | 31.6% ± 0.8% a | 4.1% ± 1.0% a | 2.9% ± 0.9% a | 7.4% ± 1.0% a | 20.6% ± 1.1% a | 27.3% ± 1.2% a | 40.1% ± 1.1% b | 53.8% ± 0.7% b |
| AT-LP | 32.6% ± 1.3% a | 11.6% ± 0.9% b | 7.7% ± 0.9% b | 7.5% ± 1.2% a | 27.4% ± 0.9% b | 30.6% ± 0.5% c | 53.6% ± 1.1% c | 73.7% ± 1.1% d |
| AT-LP-HPH | 52.3% ± 1.4% c | 15.7% ± 1.1% c | 11.8% ± 1.0% c | 15.8% ± 1.9% b | 31.7% ± 1.4% c | 42.6% ± 1.3% d | 61.4% ± 1.2% d | 91.7% ± 1.0% e |
| Protein Samples | H0-ANS |
|---|---|
| OPI | 402.08 ± 21.02 b |
| AT | 469.60 ± 18.59 c |
| AT-HPH | 563.04 ± 14.59 d |
| AT-LP | 314.92 ± 8.64 a |
| AT-LP-HPH | 335.40 ± 4.61 a |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.; Cha, Y.; Wu, F.; Xu, X.; Du, M. Effects of Limited Hydrolysis and High-Pressure Homogenization on Functional Properties of Oyster Protein Isolates. Molecules 2018, 23, 729. https://doi.org/10.3390/molecules23040729
Yu C, Cha Y, Wu F, Xu X, Du M. Effects of Limited Hydrolysis and High-Pressure Homogenization on Functional Properties of Oyster Protein Isolates. Molecules. 2018; 23(4):729. https://doi.org/10.3390/molecules23040729
Chicago/Turabian StyleYu, Cuiping, Yue Cha, Fan Wu, Xianbing Xu, and Ming Du. 2018. "Effects of Limited Hydrolysis and High-Pressure Homogenization on Functional Properties of Oyster Protein Isolates" Molecules 23, no. 4: 729. https://doi.org/10.3390/molecules23040729




