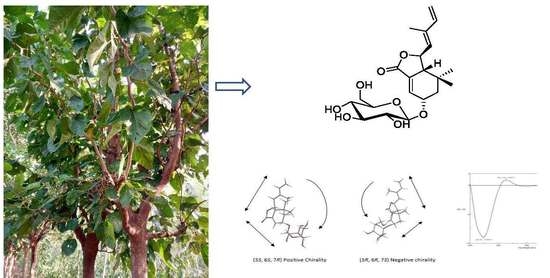
A Chemical Investigation of the Leaves of Morus alba L.
Abstract
:1. Introduction
2. Results and Discussion
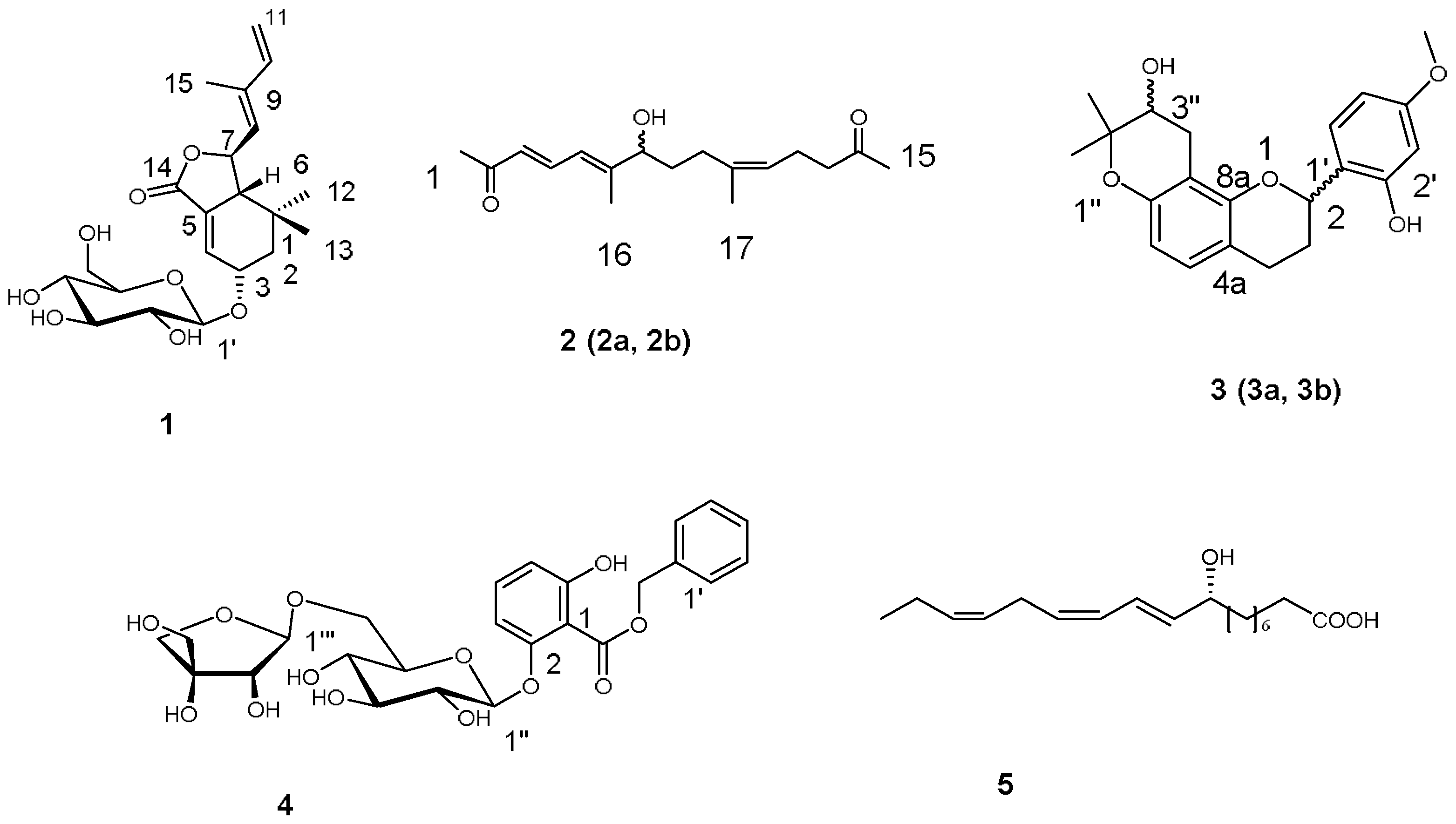
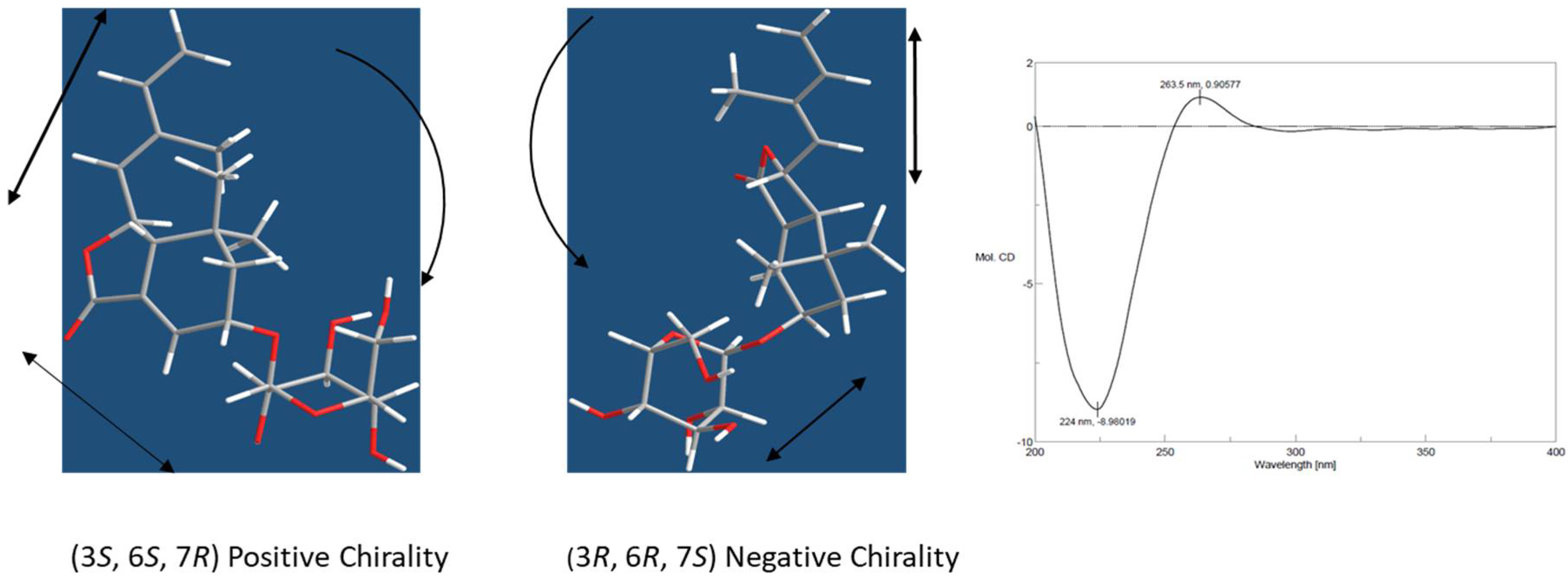
2.1. Characterization
2.2. Aldose Reductase Inhibitory Effects of 2 and 5 and Neuroprotective Effects of Compounds 1–5
2.3. Discussion
3. Materials and Methods
3.1. Plant Material
3.2. General Experimental Procedures
3.3. Cell Lines, Chemicals and Biochemicals
3.4. Extraction and Isolation
3.5. Characterization
3.6. Acid Hydrolysis of the Saponins and Determination of the Absolute Configuration of the Monosaccharides
3.7. Aldose Reductase Assay
3.8. Neuroprotection Bioassays
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Wu, Z.Y.; Raven, P.H.; Hong, D.Y. Flora of China, 2003 ed.; Science Press: Beijing, China, 2003; Missouri Botanical Garden Press: St. Louis, MI, USA, 2003; Volume 5, pp. 22–26. [Google Scholar]
- Gao, L.; Li, Y.D.; Zhu, B.K.; Li, Z.Y.; Huang, L.B.; Li, X.Y.; Wang, F.; Ren, F.C.; Liao, T.G. Two new prenylflavonoids from Morus alba. J. Asian Nat. Prod. Res. 2017, 23, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2010; Volume 1, pp. 279–281. [Google Scholar]
- Pel, P.; Chae, H.S.; Nhoek, P.; Kim, Y.M.; Chin, Y.W. Chemical Constituents with Proprotein Convertase Subtilisin/Kexin Type 9 mRNA Expression Inhibitory Activity from Dried Immature Morus alba Fruits. J. Agric. Food Chem. 2017, 65, 5316–5321. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Z.; Wang, Y.C.; Wang, Y.; Zhang, Y. Bioassay-guided screening and isolation of α-glucosidase and tyrosinase inhibitors from leaves of Morus alba. F. Food Chem. 2012, 131, 617–625. [Google Scholar] [CrossRef]
- Zhao, G.J.; Xi, Z.X.; Chen, W.X.; Li, X.; Sun, L.; Sun, L.N. Chemical constituents from Tithonia diversifolia and their chemotaxonomic significance. Biochem. Syst. Ecol. 2012, 44, 250–254. [Google Scholar] [CrossRef]
- Li, H.X.; Jo, E.; Myung, C.S.; Kim, Y.H.; Yang, S.Y. Lipolytic effect of compounds isolated from leaves of mulberry (Morus alba L.) in 3T3-L1 adipocytes. Nat. Prod. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tan, Y.X.; Chen, R.Y.; Kang, J. The latest review on the polyphenols and their bioactivities of Chinese Morus plants. J. Asian Nat. Prod. Res. 2014, 16, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Sangshetti, J.; Chouthe, R.; Sakle, N.; Gonjari, I.; Shinde, D. Aldose Reductase: A Multi-disease Target. Curr. Enzym. Inhib. 2014, 10, 2–12. [Google Scholar] [CrossRef]
- Shen, B.; Vetri, F.; Mao, L.; Xu, H.L.; Paisansathan, C.; Pelligrino, DA. Aldose reductase inhibition ameliorates the detrimental effect of estrogen replacement therapy on neuropathology in diabetic rats subjected to transient forebrain ischemia. Brain Res. 2010, 1342, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Goldin, A.; Beckman, J.A.; Schmidt, AM.; Creager, MA. Advanced Glycation End Products Sparking the Development of Diabetic Vascular Injury. Circulation 2006, 114, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C. Advanced glycation end products. Contrib. Nephrol. 2011, 170, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, H.; Nakaigawa, N.; Miyoshi, Y.; Fujinami, K.; Kubota, Y.; Uemura, H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate 2005, 64, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Kuniyasu, H.; Oue, N.; Wakikawa, A.; Shigeishi, H.; Matsutani, N.; Kuraoka, K.; Ito, R.; Yokozaki, H.; Yasui, W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J. Pathol. 2002, 196, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Ramana, K.V. ALDOSE REDUCTASE: New Insights for an Old Enzyme. Biomol. Concepts 2011, 2, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.S.; Yadav, U.; Reddy, A.; Saxena, A.; Tammali, R.; Mohammad, S.; Ansari, H.N.; Bhatnagar, A.; Petrash, J.M.; Srivastava, S.; et al. Aldose reductase inhibition suppresses oxidative stress-induced inflammatory disorders. Chem. Biol. Interact. 2011, 191, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Nakanishi, K. Exciton chirality method and its application to configurational and conformational studies of natural products. Acc. Chem. Res. 1972, 5, 257–263. [Google Scholar] [CrossRef]
- Kitagawa, I.; Hori, K.; Sakagami, M. Saponin and Sapogenol. XLIX. On the Constitutents of the Roots of Glycyrrhiza inflata BATALIN from Xinjiang, China. Characterization of Two Sweet Oleanane-Type Triterpene Oligoglycosides, Apioglycyrrhizin and Araboglycyrrhizin. Chem. Pharm. Bull. 1993, 41, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- McLean, S.F.; Reynolds, W.; Tinto, W.F.; Chan, W.R.; Shepherd, V. Complete 13C and 1H Spectral Assignments of Prenylated Flavonoids and a Hydroxy Fatty Acid from the Leaves of Caribbean Artocarpus communis. Magn. Reson. Chem. 1996, 34, 719–722. [Google Scholar] [CrossRef]
- Ramirez, M.A.; Borja, N.L. Epalrestat: An aldose reductase inhibitor for the treatment of diabetic neuropathy. Pharmacotherapy 2008, 28, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Yoon, I.S.; Au, Q.; Barber, J.R.; Ng, S.C.; Zhang, B. Development of a high-throughput screening assay for cytoprotective agents in rotenone-induced cell death. Anal. Biochem. 2010, 407, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Bansol, P.K.; Deshmukh, R. Animal Models of Neurological Disordors; Springer Nature: Singapore, 2017; p. 30. ISBN 978-981-10-5980-3. [Google Scholar]
- Javed, H.; Azimullah, S.; Abul Khair, S.B.; Ojha, S.; Haque, M.E. Neuroprotective effect of nerolidol against neuroinflammation and oxidative stress induced by rotenone. BMC Neurosci. 2016, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, M.; Rajasankar, S.; Gobi, V.V.; Dhanalakshmi, C.; Manivasagam, T.; Justin Thenmozhi, A.; Essa, M.M.; Kalandar, A.; Chidambaram, R. Neuroprotective effect of Demethoxycurcumin, a natural derivative of Curcumin on rotenone induced neurotoxicity in SH-SY 5Y Neuroblastoma cells. BMC Complement. Altern. Med. 2017, 17, 217. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Zhang, Q.J.; Zheng, Z.F.; Chen, R.Y.; Yu, D.Q. 2-Arylbenzofuran Derivatives from Morus cathayana. J. Nat. Prod. 2009, 72, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.J.; Wu, Y.; Wang, Y.H.; He, W.Y.; Chen, R.Y.; Yu, D.Q. New Diels-Alder type adducts from Morus macroura and their anti-oxidant activities. Chem. Pharm. Bull. 2004, 52, 1190–1193. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.H.; Kim, J.E.; Rhie, S.J.; Yoon, S. The Role of Oxidative Stress in Neurodegenerative Diseases. Exp. Neurobiol. 2015, 24, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Hao, Z.Y.; Zhang, G.J.; Zhang, Q.J.; Chen, R.Y.; Yu, D.Q. Cytotoxic Triterpenoid Saponins from Lysimachia clethroides. J. Nat. Prod. 2011, 74, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Tang, Y.; Liu, Q.; Guo, N.; Zhang, J.; Xiao, Z.; Chen, R.; Shen, Z. Isolation, modification, and aldose reductase inhibitory activity of rosmarinic acid derivatives from the roots of Salvia grandifolia. Fitoterapia 2016, 112, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, Y.N.; Song, X.Y.; Shao, S.Y.; Feng, Z.M.; Jiang, J.S.; Li, L.; Chen, N.H.; Zhang, P.C. Forsythoneosides A-D, Neuroprotective Phenethanoid and Flavone Glycoside Heterodimers from the Fruits of Forsythia suspensa. J. Nat. Prod. 2015, 78, 2390–2397. [Google Scholar] [CrossRef] [PubMed]
- Ercisli, S.; Orhan, E. Chemical composition of white (Morus alba), red (Morus rubra) and black (Morus nigra) mulberry fruits. Food Chem. 2007, 103, 1380–1384. [Google Scholar] [CrossRef]
- Butta, M.S.; NazirbM, A.; Sultana, M.T.; Schroën, K. Morus alba L. nature’s functional tonic. Trends Food Sci. Technol. 2008, 19, 505–512. [Google Scholar] [CrossRef]
- Arabshahi-Delouee, S.; Urooj, A. Antioxidant properties of various solvent extracts of mulberry (Morus indica L.) leaves. Food Chem. 2007, 102, 1233–1240. [Google Scholar] [CrossRef]
- Chang, L.W.; Juang, L.J.; Wang, B.S.; Wang, M.Y.; Tai, H.M.; Hung, W.J.; Chen, Y.J.; Huang, M.H. Antioxidant and antityrosinase activity of mulberry (Morus alba L.) twigs and root bark. Food Chem. Toxicol. 2011, 49, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Singab, A.N.; El-Beshbishy, H.A.; Yonekawa, M.; Nomura, T.; Fukai, T. Hypoglycemic effect of Egyptian Morus alba root bark extract: Effect on diabetes and lipid peroxidation of streptozotocin-induced diabetic rats. J. Ethnopharmacol. 2005, 100, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Kojima, T.; Makino, M.; Kimura, Y.; Fujimoto, Y. Studies on the Constituents of the Leaves of Morus alba L. Chem Pharm Bull. 2001, 49, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Oku, T.; Yamada, M.; Nakamura, M.; Sadamori, N.; Nakamura, S. Br Inhibitory effects of extractives from leaves of Morus alba on human and rat small intestinal disaccharidase activity. J. Nutr. 2006, 95, 933–938. [Google Scholar] [CrossRef]
Sample Availability: Sample of the compound 5 is available from the authors. |
| 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|
| Position | δH (J in Hz) | Position | δH (J in Hz) | Position | δH (J in Hz) | Position | δH (J in Hz) |
| 2α | 1.86, (overlapped) | 1 | 2.25, s | 2 | 5.27, dd, (9.6, 1.8) | 3 | 6.65, d, (8.4) |
| 2β | 1.60, dd, (15.0, 6.5) | 3 | 6.09, d, (16.5) | 3a | 2.20, m | 4 | 7.18, dd, (8.4) |
| 3 | 4.41, m | 4 | 7.45, dd, (16.5, 11.5) | 3b | 1.85, m | 5 | 6.55, d, (8.4) |
| 4 | 6.82, t, (3.0) | 5 | 6.23, d, (11.5) | 4a | 2.86, m | 2′, 6′ | 7.47, m |
| 6 | 2.57, m | 7 | 3.93, t, (6.5) | 4b | 2.63, m | 3′, 5′ | 7.36, m |
| 7 | 5.12, t, (9.3) | 8 | 1.47, m | 5 | 6.79, d, (8.4) | 4′ | 7.36, m |
| 8 | 5.66, d, (9.3) | 9 | 1.97, t, (7.5) | 6 | 6.30, d, (8.4) | 7′a | 5.33, d, (12.6) |
| 10 | 6.44, dd, (17.5, 9.0) | 11 | 5.03, t, (7.0) | 3′ | 6.40, d, (2.4) | 7′b | 5.22, d, (12.6) |
| 11a | 5.37, d, (17.5) | 12 | 2.12, dd, (7.5, 7.0) | 5′ | 6.42, dd, (8.4, 2.4) | 1″ | 4.84, d, (7.2) |
| 11b | 5.20, d, (10.5) | 13 | 2.42, t, (7.5) | 6′ | 7.25, d, (8.4) | 2″ | 3.21, m |
| 12 | 0.90, s | 15 | 1.06, s | 3′′ | 3.73, dd, (7.2, 6.0) | 3″ | 3.49, m |
| 13 | 0.87, 3H, s | 16 | 1.85 (3H, s) | 4′′a | 2.92, dd, (17.4, 7.8) | 4″ | 3.09, t, (8.7) |
| 15 | 1.88, s | 17 | 1.62 (3H, s) | 4′′b | 2.54, dd, (17.4, 5.4) | 5″ | 3.25, m |
| 1′ | 4.35, d, (8.0) | 5′′ | 1.22, s b | 6″a | 3.86, d, (8.1) | ||
| 2′ | 2.91, t, (8.0) | 6′′ | 1.31, s b | 6″b | 3.44, m | ||
| 3′ | 3.15, m | -OMe | 3.75, s | 1′′′ | 4.83, d, (3.0) | ||
| 4′ | 3.03, t, (8.0) | 2′′′ | 3.75, d, (3.0) | ||||
| 5′ | 3.15, m | 4′′′a | 3.89 (1H, d, 9.0) | ||||
| 6′a | 3.68, dd, (10.5, 1.0) | 4′′′a | 3.59 (1H. d, 9.0) | ||||
| 6′b | 3.44, dd, (10.5, 6.0) | 5′′′ | 3.35 (1H, m) | ||||
| 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|
| Position | δc, Type | Position | δc, Type | Position | δc, Type | Position | δc, Type |
| 1 | 29.5, C | 1 | 27.1, CH3 | 2 | 74.2, CH | 1 | 120.0, C |
| 2 | 43.2, CH2 | 2 | 198.3, CH | 3 | 29.9, CH2 | 2 | 155.3, C |
| 3 | 70.4, CH | 3 | 129.9, CH | 4 | 25.8, CH2 | 3 | 105.5 b, CH |
| 4 | 132.4, CH | 4 | 139.4, CH | 5 | 128.5, CH | 4 | 131.0, CH |
| 5 | 130.7, C | 5 | 122.3, CH | 6 | 109.7, CH | 5 | 109.4 b, CH |
| 6 | 52.3, CH | 6 | 153.1, C | 7 | 153.0, C | 6 | 155.4, C |
| 7 | 76.8, CH | 7 | 74.7, CH | 8 | 109.3, C | 7 | 165.8, C |
| 8 | 129.8, CH | 8 | 33.3, CH2 | 4a | 114.4, C | 1′ | 136.2, C |
| 9 | 138.8, C | 9 | 27.5, CH2 | 8a | 154.5, C | 2′, 6′ | 127.8, CH |
| 10 | 140.0, CH | 10 | 135.3, C | 1′ | 122.4, C | 3′, 5′ | 128.3, CH |
| 11 | 115.6, CH2 | 11 | 124.0, CH | 2′ | 156.1, C | 4′ | 127.8, CH |
| 12 | 21.0, CH3 | 12 | 21.8, CH2 | 3′ | 102.1, CH | 7′ | 66.0, CH2 |
| 13 | 28.9, CH3 | 13 | 43.1, CH2 | 4′ | 161.4, C | 1” | 100.4, CH |
| 14 | 168.8, C | 14 | 208.1, C | 5′ | 105.7, CH | 2” | 73.27, CH |
| 15 | 12.3, CH3 | 15 | 29.8, CH3 | 6′ | 128.0, CH | 3” | 75.6, CH |
| 1′ | 102.7, CH | 16 | 13.4, CH3 | 2” | 77.4, C | 4” | 70.0, CH |
| 2′ | 73.5, CH | 17 | 23.2, CH3 | 3” | 70.5, CH | 5” | 76.8, CH |
| 3′ | 76.8, CH | 4” | 27.3, CH2 | 6” | 67.8, CH2 | ||
| 4′ | 70.1, CH | 5′′ | 20.8 | 1′′′ | 109.4, CH | ||
| 5′ | 76.9, CH | 6′′ | 25.8 | 2′” | 75.9, CH | ||
| 6′ | 61.2, CH2 b 4.29, dd, (11.5, 6) | -OMe | 55.7 | 3′′′ | 78.7, C | ||
| 4′′′ | 73.30, CH2 | ||||||
| 5′′′ | 63.2, CH2 | ||||||
| Sample | Serum Deprivation (%) | Nicouline 4 μM (%) |
|---|---|---|
| control | 100.0 ± 3.7 | 100.0 ± 1.4 |
| model | 41.4 ± 3.8 ### | 74.3 ± 1.4 ### |
| 1 | 50.2 ± 12.7 | 78.6 ± 2.9 |
| 2 | 66.2 ± 12.6 | 78.4 ± 2.0 * |
| 3 | 59.8 ± 2.7*** | 75.7 ± 3.0 |
| 4 | 50.9 ± 7.8 | 76.9 ± 3.6 |
| 5 | 70.2 ± 16.1 | 86.2 ± 7.6 *** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.-y.; Zhang, T.; Wang, X.; Hamann, M.T.; Kang, J.; Yu, D.-q.; Chen, R.-y. A Chemical Investigation of the Leaves of Morus alba L. Molecules 2018, 23, 1018. https://doi.org/10.3390/molecules23051018
Chen X-y, Zhang T, Wang X, Hamann MT, Kang J, Yu D-q, Chen R-y. A Chemical Investigation of the Leaves of Morus alba L. Molecules. 2018; 23(5):1018. https://doi.org/10.3390/molecules23051018
Chicago/Turabian StyleChen, Xiao-yan, Ting Zhang, Xin Wang, Mark T. Hamann, Jie Kang, De-quan Yu, and Ruo-yun Chen. 2018. "A Chemical Investigation of the Leaves of Morus alba L." Molecules 23, no. 5: 1018. https://doi.org/10.3390/molecules23051018
APA StyleChen, X.-y., Zhang, T., Wang, X., Hamann, M. T., Kang, J., Yu, D.-q., & Chen, R.-y. (2018). A Chemical Investigation of the Leaves of Morus alba L. Molecules, 23(5), 1018. https://doi.org/10.3390/molecules23051018