Chemical Constituents from Cimicifuga dahurica and Their Anti-Proliferative Effects on MCF-7 Breast Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of Compounds 1–17
2.2. Cytotoxicity Activity
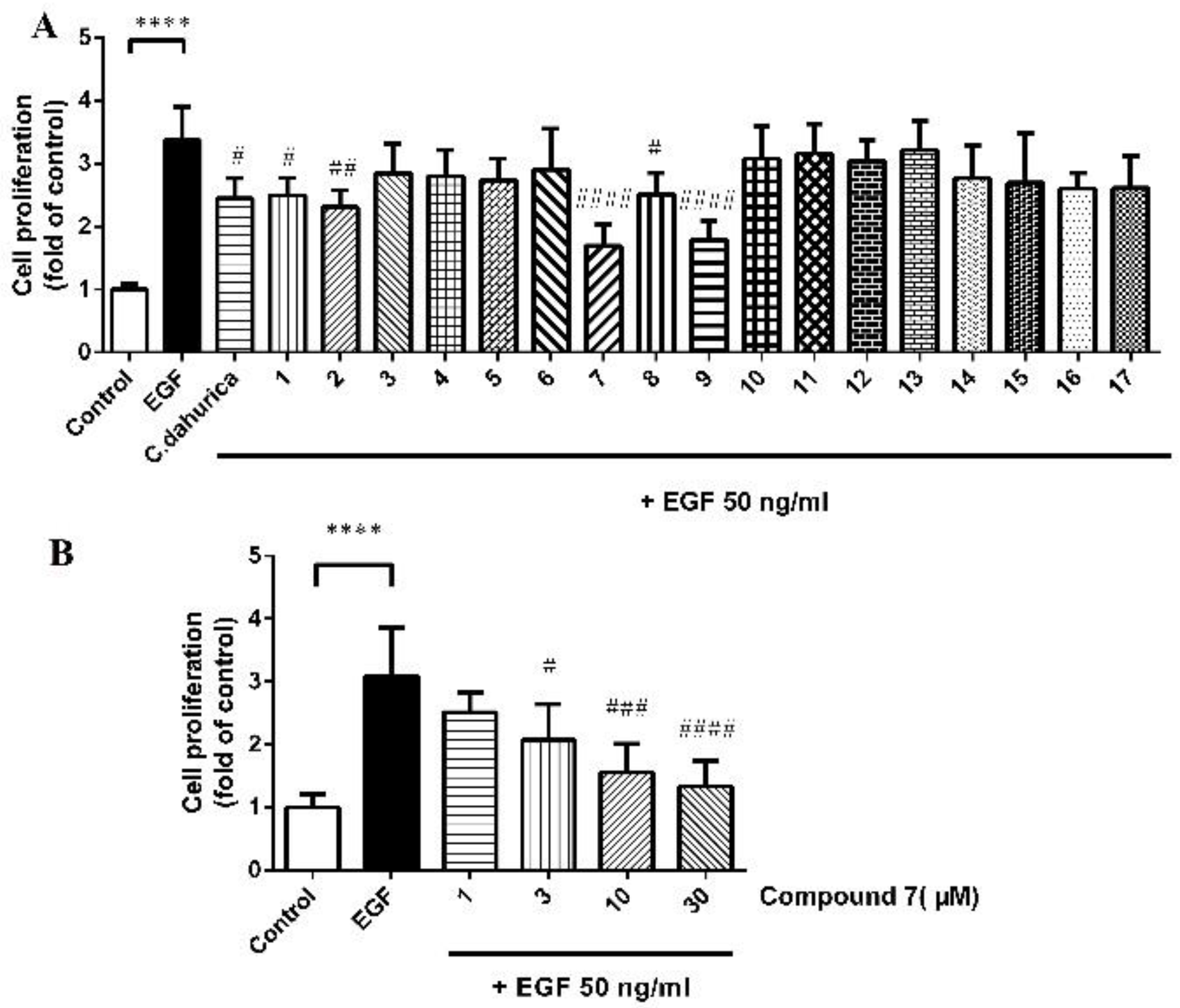
2.2.1. Cell Proliferation Activity
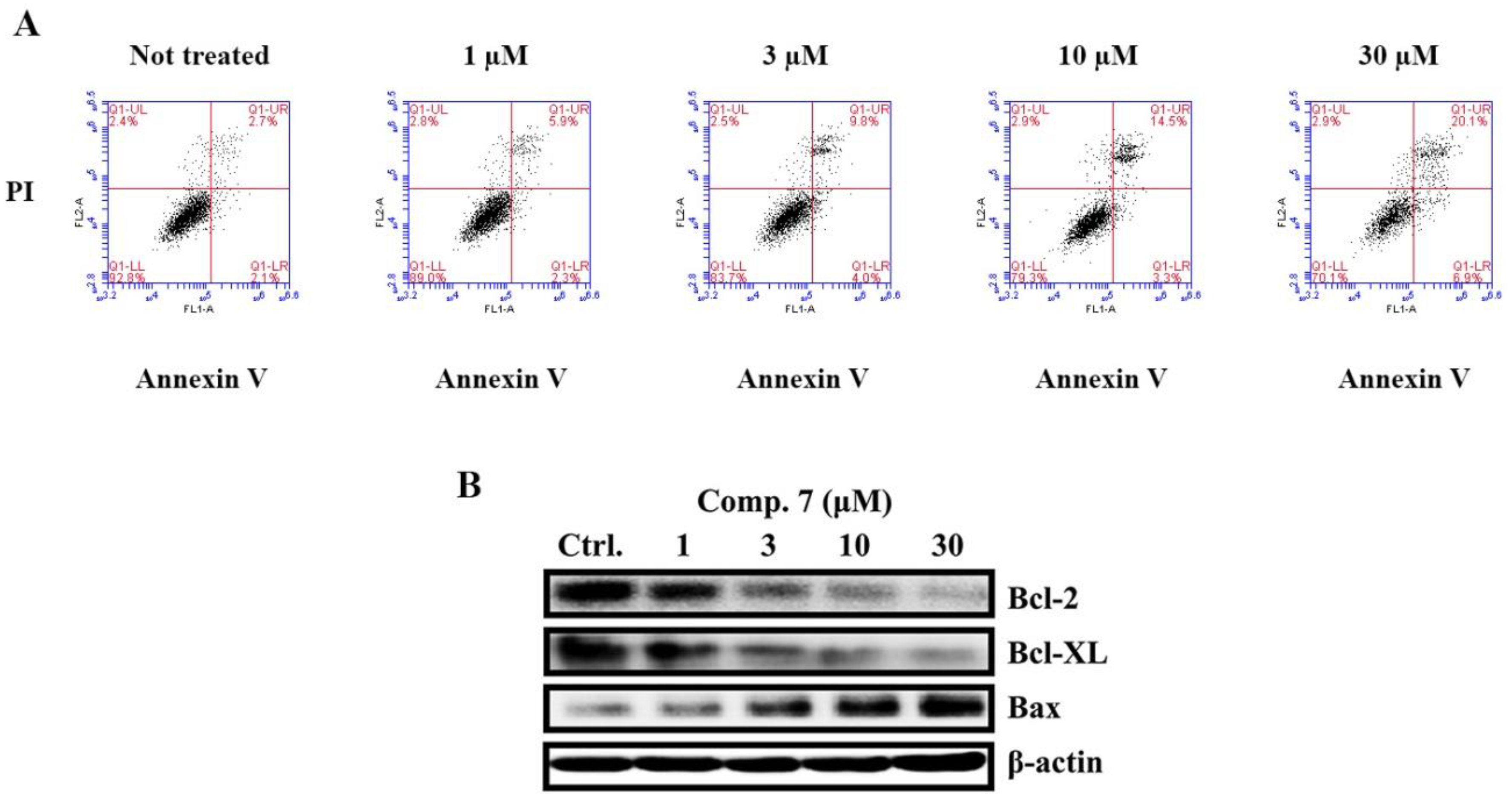
2.2.2. Cell Cycle Distribution and Western Blot Analyses
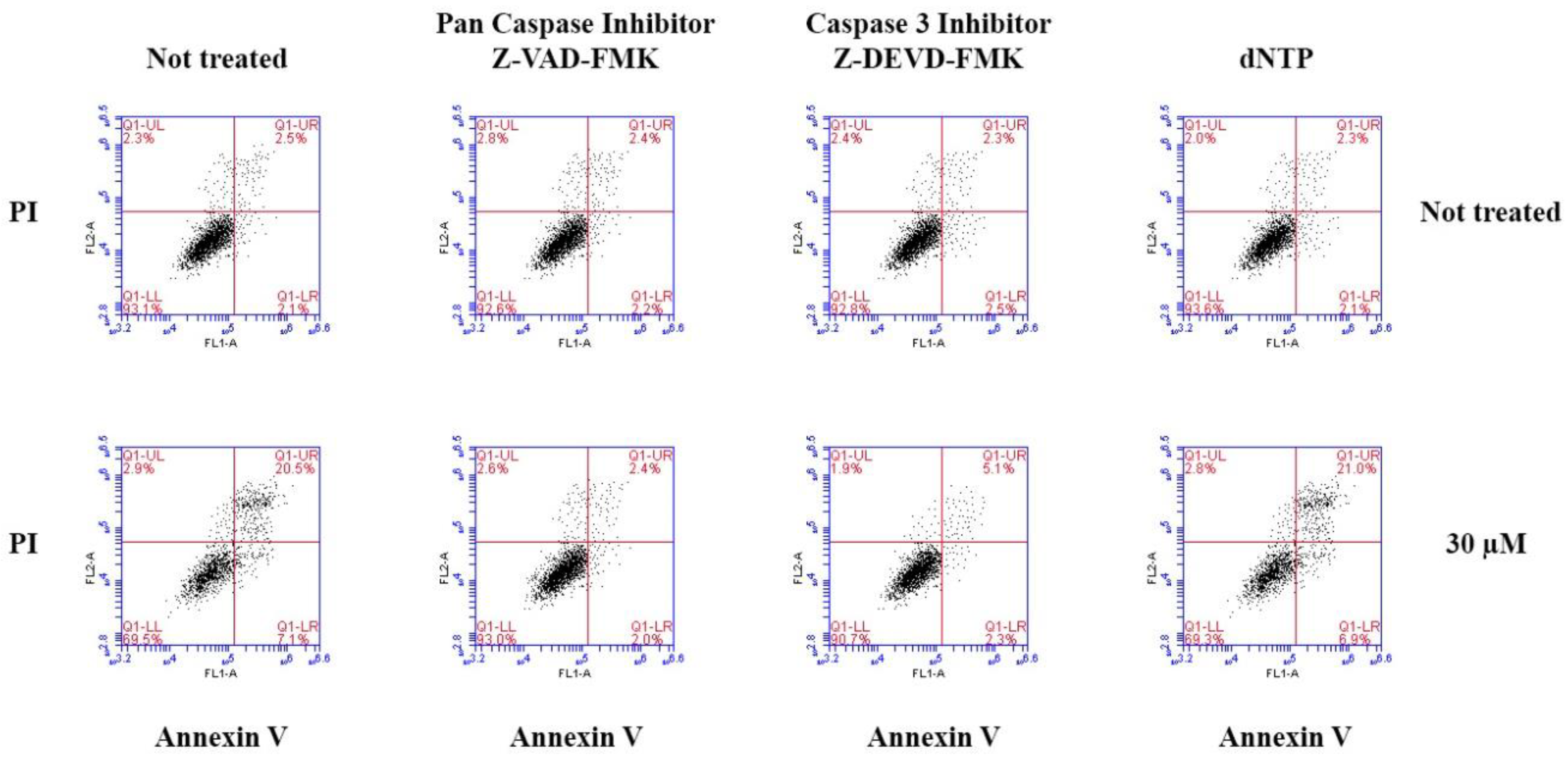
2.2.3. Induction of Apoptosis by Compound 7 in MCF-7 Cells
3. Materials and Methods
3.1. General Experimental Proceduces
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Acid hydrolysis and Sugar Identification
3.5. Biological Evaluations
3.5.1. Cell Culture
3.5.2. Anti-Proliferative Activity
3.5.3. Cell Cycle Distribution Analyses
3.5.4. Annexin V/PI Analyses
3.5.5. Western Blotting
3.5.6. Statistics
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Guo, Y.Q.; Yin, T.; Wang, X.M.; Zhang, F.; Pan, G.X.; Lv, H.; Wang, X.R.; Owoicho Orgah, J.; Zhu, Y.; Wu, H.H. Traditional uses, phytochemistry, pharmacology and toxicology of the genus Cimicifuga: A review. J. Ethnopharmacol. 2017, 209, 264–282. [Google Scholar] [CrossRef] [PubMed]
- The State Pharmacopoeia Commission of P.R.China. Pharmacopoeia of Chinese People’s Republic; Medicine Science and Technology Press of China: Beijing, China, 2015; pp. 73–74. ISBN 9787506773379. [Google Scholar]
- Li, J.X.; Yu, Z.Y. Cimicifugae rhizoma: From origins, bioactive constituents to clinical outcomes. Curr. Med. Chem. 2006, 13, 2927–2951. [Google Scholar] [PubMed]
- Hao, D.C.; Gu, X.J.; Xiao, P.G.; Liang, Z.G.; Xu, L.J.; Peng, Y. Recent advance in chemical and biological studies on cimicifugeae pharmaceutical resources. Chin. Herb. Med. 2013, 5, 81–95. [Google Scholar] [CrossRef]
- Borrelli, F.; Ernst, E. Black cohosh (Cimicifuga racemosa) for menopausal symptoms: A systematic review of its efficacy. Pharmacol. Res. 2008, 58, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Si, J.; Chang, Q.; Zhou, L.; Chen, S.; Xiao, P.; Wu, E. Antitumor activity and mechanisms of action of total glycosides from aerial part of Cimicifuga dahurica targeted against hepatoma. BMC Cancer 2007, 7, 237. [Google Scholar] [CrossRef] [PubMed]
- Schmid, D.; Woehs, F.; Svoboda, M.; Thalhammer, T.; Chiba, P.; Moeslinger, T. Aqueous extracts of Cimicifuga racemosa and phenolcarboxylic constituents inhibit production of proinflammatory cytokines in LPS-stimulated human whole blood. Can. J. Physiol. Pharmacol. 2009, 87, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Seidlova-Wuttke, D.; Stecher, G.; Kammann, M.; Haunschild, J.; Eder, N.; Stahnke, V.; Wessels, J.; Wuttke, W. Osteoprotective effects of Cimicifuga racemosa and its triterpene-saponins are responsible for reduction of bone marrow fat. Phytomedicine 2012, 19, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, F.; Ernst, E. Black cohosh (Cimicifuga racemosa): A systematic review of adverse events. Am. J. Obstet. Gynecol. 2017, 199, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Nian, Y.; Wang, H.Y.; Zhou, L.; Su, J.; Li, Y.; Qiu, M.H. Cytotoxic cycloartane triterpenes of the traditional Chinese medicine “shengma” (Cimicifuga dahurica). Planta Med. 2013, 79, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Li, P.L.; Tang, X.L.; Wang, S.J.; Jiang, Y.T.; Shen, L.; Xu, B.M.; Shao, Y.L.; Li, G.Q. Cycloartane triterpenoids and their glycosides from the rhizomes of Cimicifuga foetida. J. Nat. Prod. 2014, 77, 1997–2005. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Mimaki, Y.; Sakagami, H.; Sashida, Y. Cycloartane glycosides from the rhizomes of Cimicifuga racemosa and their cytotoxic activities. Chem. Pharm. Bull. 2002, 50, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.G.; Xie, S.; Lee, J.K.; Kwok, H.F.; Gao, S.; Nian, Y.; Wu, X.X.; Wong, C.K.; Qiu, M.H.; Lau, C.B. New potential beneficial effects of actein, a triterpene glycoside isolated from Cimicifuga species, in breast cancer treatment. Sci. Rep. 2016, 6, 35263. [Google Scholar] [CrossRef] [PubMed]
- Jarry, H.; Stromeier, S.; Wuttke, W.; Nahrstedt, A. Petasiphenone, a phenol isolated from Cimicifuga racemosa, in vitro inhibits proliferation of the human prostate cancer cell line LNCaP. Planta Med. 2007, 73, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.H.; Kim, H.J.; Park, S.H.; Kim, J.; Williams, D.R.; Jung, D.W.; Lee, I.S. Cytotoxic caffeic acid derivatives from the rhizomes of Cimicifuga heracleifolia. Arch. Pharm. Res. 2012, 35, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Deyama, T. The constituents of Eucommia ulmoides Oliv. I. Isolation of (+)-medioresinol di-O-β-d-glucopyranoside. Chem. Pharm. Bull. 1983, 31, 2993–2997. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Ye, W.C.; Hsiao, W.W.; Zhao, S.X.; Che, C.T. Cycloartane glycosides from Cimicifuga dahurica. Chem. Pharm. Bull. 2001, 49, 1468–1470. [Google Scholar] [CrossRef] [PubMed]
- Lelono, R.A.; Tachibana, S.; Itoh, K. Isolation of antifungal compounds from Gasdenia jasminoides. Pak. J. Biol. Sci. 2009, 12, 949–956. [Google Scholar] [PubMed]
- Yen, P.H.; Kiem, P.V.; Nhiem, N.X.; Tung, N.H.; Quang, T.H.; Minh, C.V.; Kim, J.W.; Choi, E.M.; Kim, Y.H. A new monoterpene glycoside from the roots of Paeonia lactiflora increases the differentiation of osteoblastic MC3T3-E1 cells. Arch. Pharm. Res. 2007, 30, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, N.; Koeda, M.; Inoue, T.; Nagai, M. Studies on the Chinese crude drug “Shoma.” VIII. Two new triterpenol bisdesmosides, 3-arabinosyl-24-O-acetylhydroshengmanol 15-glucoside and 3-xylosyl-24-O-acetylhydroshengmanol 15-glucoside, from Cimicifuga dahurica. Chem. Pharm. Bull. 1994, 42, 48–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, R.M.; Wu, S.J.; Sun, A.L. Separation and purification of four chromones from radix saposhnikoviae by high-speed counter-current chromatography. Phytochem. Anal. 2008, 19, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Li, A.F.; Xuan, H.Z.; Sun, A.L.; Liu, R.M.; Cui, J.C. Preparative separation of polyphenols from water-soluble fraction of Chinese propolis using macroporous absorptive resin coupled with preparative high performance liquid chromatography. J. Chromatogr. B 2016, 1012 1013, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Kusano, A.; Shibano, M.; Kusano, G.; Miyase, T. Studies on the constituents of Cimicifuga species. XIX. Eight new glycosides from Cimicifuga simplex Wormsk. Chem. Pharm. Bull. 1996, 44, 2078–2085. [Google Scholar] [CrossRef] [PubMed]
- Li, J.X.; Kadota, S.; Hattori, M.; Yoshimachi, S.; Shiro, M.; Oogami, N.; Mizuno, H.; Namba, T. Constituents of Cimicifugae rhizoma. I. Isolation and characterization of ten new cycloartenol triterpenes from Cimicifuga heracleifolia Komarov. Chem. Pharm. Bull. 1993, 41, 832–841. [Google Scholar] [CrossRef]
- Kusano, A.; Shibano, M.; Kitagawa, S.; Kusano, G.; Nozoe, S.; Fushiya, S. Studies on the constituents of Cimicifuga species. XV. Two new diglycosides from the aerial parts of Cimicifuga simplex Wormsk. Chem. Pharm. Bull. 1994, 42, 1940–1943. [Google Scholar] [CrossRef]
- Cuong, T.D.; Lim, C.J.; Kim, S.W.; Park, J.E.; Hung, T.M.; Min, B.S. Isolation of compounds from Cimicifugae rizhoma and their cytototoxic activity. Nat. Prod. Sci. 2011, 17, 80–84. [Google Scholar]
- Kusano, A.; Takahira, M.; Shibano, M.; Miyase, T.; Kusano, G. Studies on the constituents of Cimicifuga Species. XXVI. Twelve new cyclolanostanol glycosides from the underground parts of Cimicifuga simplex Wormsk. Chem. Pharm. Bull. 1999, 47, 511–516. [Google Scholar] [CrossRef]
- Kadota, S.; Li, J.X.; Tanaka, K.; Namba, T. Constituents of Cimicifugae rizhoma II. Isolation and structures of new cycloartenol triterpenoids and related compounds from Cimicifuga foetida L. Tetrahedron 1995, 51, 1143–1166. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Ye, W.C.; Che, C.T.; Zhao, S.X. A new cycloartane saponin from Cimicifuga acerina. J. Asian Nat. Prod. Res. 1999, 2, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Nuntanakorn, P.; Jiang, B.; Einbond, L.S.; Yang, H.; Kronenberg, F.; Weinstein, I.B.; Kennelly, E.J. Polyphenolic constituents of Actaea racemosa. J. Nat. Prod. 2006, 69, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Han, L.F.; Pan, G.X.; Peng, S.; Andre, N. A new phenolic amide glycoside from Cimicifuga dahurica. Acta Pharm. Sin. 2013, 48, 1281–1285. [Google Scholar]
- Knudsen, K.E.; Diehl, J.A.; Haiman, C.A.; Knudsen, E.S. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene 2006, 25, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Huang, H.T.; Wang, Z.X.; Zhang, G.B. The inflammatory CXC chemokines, GROαhigh, IP-10low, and MIGlow, in tumor microenvironment can be used as new indicators for non-small cell lung cancer progression. Immunol. Investig. 2017, 46, 361–374. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–17 are available from the authors. |



| Position | δCa | δHb (Mult., J in Hz) | Position | δCa | δHb (Mult., J in Hz) |
|---|---|---|---|---|---|
| 1 | 130.9 | - | 5′ | 116.2 | 6.75 (d, 8.0) |
| 2 | 112.4 | 7.17 * | 6′ | 122.2 | 6.68 (dd, 1.5, 8.0) |
| 3 | 150.9 | - | 7′ | 36.1 | 2.79 (t, 7.5) |
| 4 | 149.7 | - | 8′ | 42.4 | 3.51 (t, 7.5) |
| 5 | 117.3 | 7.17 * | 3-OCH3 | 56.7 | 3.88 (s) |
| 6 | 122.7 | 7.12 (br s) | 3′-OCH3 | 56.4 | 3.84 (s) |
| 7 | 141.4 | 7.47 (d, 15.5) | 1″ | 100.2 | 5.32 (d, 8.0) |
| 8 | 120.4 | 6.50 (d, 15.5) | 2″ | 72.0 | 3.70 * |
| 9 | 168.8 | - | 3″ | 72.9 | 4.18 (t, 3.0) |
| 1′ | 132.0 | - | 4″ | 68.6 | 3.63 (dd, 3.0, 10.0) |
| 2′ | 113.5 | 6.83 (d, 1.5) | 5″ | 75.9 | 3.87 (m) |
| 3′ | 148.9 | - | 6″ | 62.8 | 3.71 * |
| 4′ | 146.0 | - | 3.88 (dd, 6.0, 12.0) |
| Position | 4 | 7 | ||
|---|---|---|---|---|
| δCa | δHb (Mult., J in Hz) | δCa | δHb (Mult., J in Hz) | |
| 1, 5 | 55.7 | 3.14 (br s) | 55.4 | 3.09 (br s) |
| 2, 6 | 87.2 | 4.79 (d, 7.5) | 87.0 | 4.74 (br s) |
| 4, 8 | 73.0 | 3.96 (dd, 3.0, 9.0) | 72.7 | 3.86 * |
| 4.32 (dd, 6.5, 9.0) | 4.22 (d, 6.0) | |||
| 1′, 1″ | 139.4 | - | 137.1 | - |
| 2′, 2″ | 104.8 | 6.74 (s) | 111.6 | 7.01 (br s) |
| 3′, 3″ | 154.4 | - | 150.8 | - |
| 4′, 4″ | 136.0 | - | 147.6 | - |
| 5′, 5″ | 154.4 | - | 117.7 | 7.16 (d, 8.5) |
| 6′, 6″ | 104.8 | 6.74 (s) | 119.8 | 6.90 (br d, 8.0) |
| OCH3 | 57.1 | 3.88 (s) | 56.8 | 3.88 (s) |
| Allo-1′″, 1″″ | 103.9 | 5.17 (d, 7.5) | 100.6 | 5.26 (d, 7.5) |
| 2′″, 2″″ | 73.1 | 3.62 * | 72.0 | 3.63 * |
| 3′″, 3″″ | 72.2 | 4.15 (t, 2.5) | 72.8 | 4.17 (br s) |
| 4′″, 4″″ | 68.7 | 3.65 * | 68.6 | 3.60 * |
| 5′″, 5″″ | 76.3 | 3.64 (m) | 75.7 | 3.82 m |
| 6′″, 6″″ | 63.0 | 3.78 (dd, 6.0, 12.0) | 62.8 | 3.65 * |
| 3.68 * | 3.85 (dd, 6.0, 12.0) | |||
| Position | δCa | δHb (Mult., J in Hz) | Position | δCa | δHb (Mult., J in Hz) |
|---|---|---|---|---|---|
| 1 | 33.5 | 1.29 (m)/1.57 (m) | 24 | 90.1 | 3.45 (s) |
| 2 | 30.5 | 1.95 (m)/1.70 (m) | 25 | 71.9 | - |
| 3 | 90.1 | 3.21 (dd, 4.5, 11.5) | 26 | 27.3 | 1.20 (s) |
| 4 | 42.0 | - | 27 | 25.3 | 1.13 (s) |
| 5 | 49.8 | 1.72 (m) | 28 | 12.6 | 1.16 (s) |
| 6 | 22.1 | 0.81 (m)/1.62 (m) | 29 | 26.0 | 1.06 (s) |
| 7 | 27.3 | 1.96 (m)/1.21 (m) | 30 | 15.5 | 0.91 (s) |
| 8 | 48.8 | 1.36 (m) | 3-Xylose | ||
| 9 | 21.2 | - | 1′ | 107.3 | 4.30 (d, 8.0) |
| 10 | 27.7 | - | 2′ | 75.6 | 3.28 (dd, 7.5, 8.0) |
| 11 | 27.0 | 1.22 (m)/2.15 (m) | 3′ | 78.1 | 3.34 (dd, 8.0, 8.0) |
| 12 | 34.8 | 1.66 (m)/1.74 (m) | 4′ | 71.3 | 3.58 * |
| 13 | 42.5 | - | 5′ | 66.7 | 3.19 (dd, 10.0, 11.0) |
| 14 | 48.3 | - | 3.84 (dd, 5.0, 11.0) | ||
| 15 | 89.0 | 3.97 (s) | 15-Glucose | ||
| 16 | 112.4 | - | 1″ | 105.2 | 4.28 (d, 7.5) |
| 17 | 60.3 | 1.46 (d, 11.0) | 2″ | 75.4 | 3.21 (dd, 8.0, 9.0) |
| 18 | 19.9 | 1.17 (s) | 3″ | 78.0 | 3.37 * |
| 19 | 32.0 | 0.43 (d, 4.0)/0.68 (d, 4.0) | 4″ | 72.4 | 3.26 * |
| 20 | 25.0 | 1.68 (m) | 5″ | 77.5 | 3.31 (m) |
| 21 | 20.2 | 0.93 (d, 6.5) | 6″ | 63.0 | 3.62 (dd, 5.5, 11.5) |
| 22 | 38.8 | 1.03 (m)/2.35 (ddd, 2.5, 6.0, 13.0) | 3.79 (dd, 2.5, 11.5) | ||
| 23 | 72.2 | 4.42 (br d, 9.0) | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huyen, C.T.T.; Luyen, B.T.T.; Khan, G.J.; Oanh, H.V.; Hung, T.M.; Li, H.-J.; Li, P. Chemical Constituents from Cimicifuga dahurica and Their Anti-Proliferative Effects on MCF-7 Breast Cancer Cells. Molecules 2018, 23, 1083. https://doi.org/10.3390/molecules23051083
Huyen CTT, Luyen BTT, Khan GJ, Oanh HV, Hung TM, Li H-J, Li P. Chemical Constituents from Cimicifuga dahurica and Their Anti-Proliferative Effects on MCF-7 Breast Cancer Cells. Molecules. 2018; 23(5):1083. https://doi.org/10.3390/molecules23051083
Chicago/Turabian StyleHuyen, Chu Thi Thanh, Bui Thi Thuy Luyen, Ghulam Jilany Khan, Ha Van Oanh, Ta Manh Hung, Hui-Jun Li, and Ping Li. 2018. "Chemical Constituents from Cimicifuga dahurica and Their Anti-Proliferative Effects on MCF-7 Breast Cancer Cells" Molecules 23, no. 5: 1083. https://doi.org/10.3390/molecules23051083
APA StyleHuyen, C. T. T., Luyen, B. T. T., Khan, G. J., Oanh, H. V., Hung, T. M., Li, H.-J., & Li, P. (2018). Chemical Constituents from Cimicifuga dahurica and Their Anti-Proliferative Effects on MCF-7 Breast Cancer Cells. Molecules, 23(5), 1083. https://doi.org/10.3390/molecules23051083






