Salvia castanea Hairy Roots are More Tolerant to Phosphate Deficiency than Salvia miltiorrhiza Hairy Roots Based on the Secondary Metabolism and Antioxidant Defenses
Abstract
:1. Introduction
2. Results and Discussion
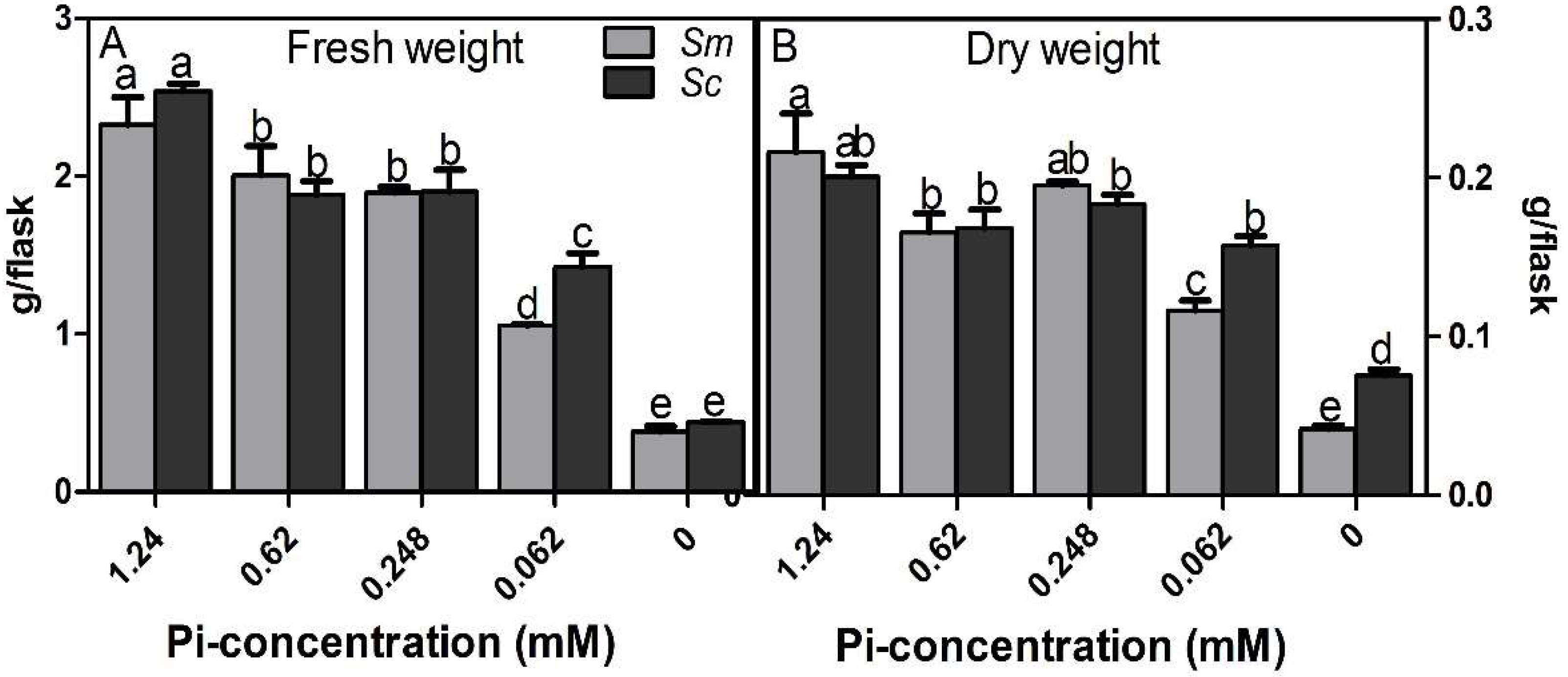
2.1. Biomass of Hairy Roots under Different Pi Concentrations
2.2. Effects of Pi Deficiency on the Content and Yields of Specialized Metabolites in Hairy Roots
2.3. Effects of Pi Deficiency on the Quantification of the Transcript Levels of Key Enzyme Genes in Hairy Roots
2.4. Expression Response of the SPX Genes to Pi Deficiency
2.5. Pi Accumulation in Hairy Roots at Control and Pi Deficiency Conditions
2.6. Antioxidant Enzyme Responses to Pi Deficiency
2.7. Effects of Pi Deficiency on the Antioxidant Activity of Salvia Hairy Roots
3. Materials and Methods
3.1. Plant Material and Pi Deficiency Treatment
3.2. High-Performance Liquid Chromatography (HPLC)
3.3. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (qRT-PCR) Assays
3.4. Measurement of Effective Pi Concentration
3.5. Analysis of Enzyme Activities
3.6. DPPH and ABTS+ Radical Cation Screening Assay
3.7. Data Analysis
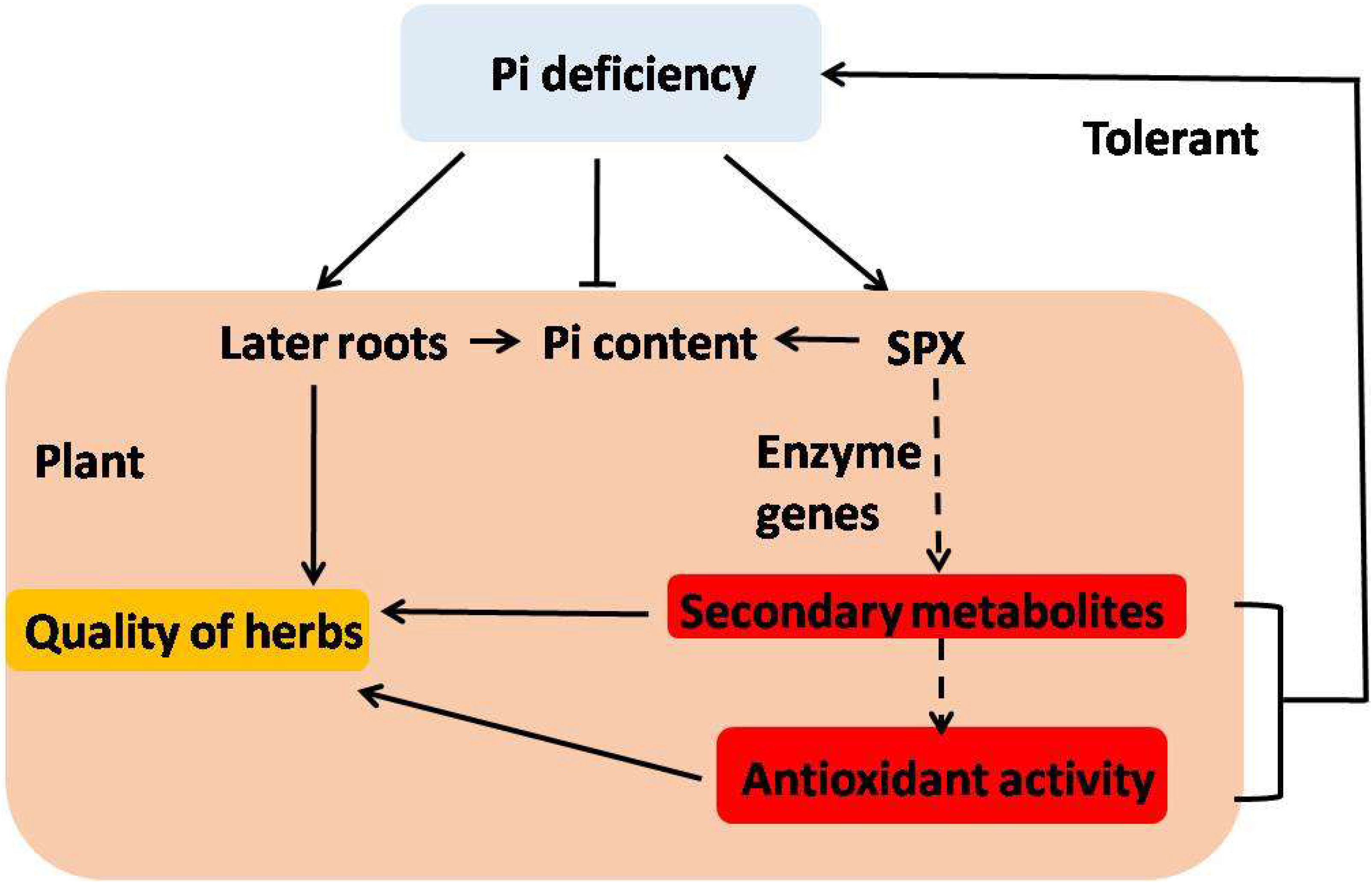
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lópezarredondo, D.L.; Leyvagonzález, M.A.; Gonzálezmorales, S.I.; Lópezbucio, J.; Herreraestrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Mehra, P.; Pandey, B.K.; Giri, J. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol. J. 2017, 15, 1054–1067. [Google Scholar] [CrossRef] [PubMed]
- Chorpet, S.; Nadia, B.; Mushtak, K.; Chanakan, P.U.T.; Patrick, D.; Hatem, R. The Involvement of OsPHO1;1 in the Regulation of Iron Transport Through Integration of Phosphate and Zinc Deficiency Signaling. Front. Plant Sci. 2016, 7, 396. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, L.; Han, G.; Cheng, B. Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Rep. 2018, 37, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Lin, Y.; Ren, J.; Bai, L.; Miao, Y.; An, G.; Song, C. Modulation of the Phosphate-Deficient Responses by the MicroRNA156 and its Targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol. 2016, 57, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liao, H.; Lucas, W.J. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. Mol. Plant 2014, 56, 192–220. [Google Scholar] [CrossRef] [PubMed]
- Nanamori, M.; Shinano, T.; Wasaki, J.; Yamamura, T.; Rao, I.M.; Osaki, M. Low Phosphorus Tolerance Mechanisms: Phosphorus Recycling and Photosynthate Partitioning in the Tropical Forage Grass, Brachiaria Hybrid Cultivar Mulato Compared with Rice. Plant Cell Physiol. 2004, 45, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; Broadley, M.R.; White, P.J. Genetic responses to phosphorus deficiency. Ann. Bot. 2004, 94, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Müller, R. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 2007, 143, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate Starvation Responses and Gibberellic Acid Biosynthesis Are Regulated by the MYB62 Transcription Factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.K.; Mehra, P.; Verma, L.; Bhadouria, J.; Giri, J. OsHAD1, a haloacid dehalogenase-like APase enhances phosphate accumulation. Plant Physiol. 2017, 174, 2316–2332. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Liang, Z.; Han, R.; Dong, J.e. Growth and saikosaponin production of the medicinal herb Bupleurum chinense DC. under different levels of nitrogen and phosphorus. Ind. Crops Prod. 2009, 29, 96–101. [Google Scholar] [CrossRef]
- Chen, H.; Chen, F.; Zhang, Y.-L.; Song, J.-Y. Production of lithospermic acid B and rosmarinic acid in hairy root cultures of Salvia miltiorrhiza. J. Ind. Microbiol. Biotechnol. 1999, 22, 133–138. [Google Scholar] [CrossRef]
- Rong, Q.; Jiang, D.; Chen, Y.; Shen, Y.; Yuan, Q.; Lin, H.; Zha, L.; Zhang, Y.; Huang, L. Molecular Cloning and Functional Analysis of Squalene Synthase 2(SQS2) in Salvia miltiorrhiza Bunge. Front. Plant. Sci. 2016, 7, 1274. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, C.; Li, C.; Zhao, Q.; Liu, L.; Fang, X.; Chen, X.Y. Recent advances in biosynthesis of bioactive compounds in traditional Chinese medicinal plants. Sci. Bull. 2016, 61, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, C.; Peng, L.; Liang, Z.; Yan, X.; Zhu, Y.; Liu, Y. Comparison of essential oil composition and phenolic acid content of selected Salvia species measured by GC–MS and HPLC methods. Ind. Crops Prod. 2015, 69, 329–334. [Google Scholar] [CrossRef]
- Jia, Y.; Bai, Z.; Pei, T.; Ding, K.; Liang, Z.; Gong, Y. The Protein Kinase SmSnRK2.6 Positively Regulates Phenolic Acid Biosynthesis in Salvia miltiorrhiza by Interacting with SmAREB1. Front. Plant Sci. 2017, 8, 1384. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Xu, H.; Zhou, C.; Liao, P.; Xiao, J.; Luo, X.; You, L.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, P.; Yang, D.; Li, W.; Liang, Z.; Liu, Y.; Liu, F. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS ONE 2013, 8, e73259. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Liang, T.; Zhang, H.; He, Z.; Liang, Z. Phosphate starvation promoted the accumulation of phenolic acids by inducing the key enzyme genes in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2016, 35, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Zhong, Y.; Wang, Y.; Wang, Z.; Li, Z.; Jing, S.; Wu, Z.; Yu, L.; Mao, C.; Yi, K. SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice. Plant Cell 2014, 26, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Yin, D.; Zhao, X. Influence of Ecological Factors on the Production of Active Substances in the anti-cancer plant Sinopodophyllum hexandrum (Royle) T.S. Ying. PLoS ONE 2015, 10, e0122981. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, F.; Lv, Z.; Li, D. Phytochemical composition, antioxidant activity and HPLC fingerprinting profiles of three Pyrola species from different regions. PLoS ONE 2014, 9, e96329. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, D.; Li, N.; Hou, X.; Wang, D.; Li, D.; Liu, J. Influence of Environmental Factors on the Active Substance Production and Antioxidant Activity in Potentilla fruticosa L. and Its Quality Assessment. Sci. Rep. 2016, 6, 28591. [Google Scholar] [CrossRef] [PubMed]
- Matkowski, A.; Zielinska, S.; Oszmianski, J.; Lamer-Zarawska, E. Antioxidant activity of extracts from leaves and roots of Salvia miltiorrhiza Bunge, S. przewalskii Maxim., and S. verticillata L. Bioresour. Technol. 2008, 99, 7892–7896. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, B.; Li, H.; Peng, L.; Ru, M.; Liang, Z.; Yan, X.; Zhu, Y. Establishment of Salvia castanea Diels f. tomentosa Stib. hairy root cultures and the promotion of tanshinone accumulation and gene expression with Ag+, methyl jasmonate, and yeast extract elicitation. Protoplasma 2015, 253, 87–100. [Google Scholar] [PubMed]
- Manisha, N.; Raghavendrarao, S.; Vandna, R.; Ajay, J. Deciphering Phosphate Deficiency-Mediated Temporal Effects on Different Root Traits in Rice Grown in a Modified Hydroponic System. Front. Plant Sci. 2016, 7, 550. [Google Scholar] [CrossRef]
- Yang, D.; Ma, P.; Liang, X.; Liang, Z.; Zhang, M.; Shen, S.; Liu, H.; Liu, Y. Metabolic Profiles and cDNA-AFLP Analysis of Salvia miltiorrhiza and Salvia castanea Diel f. tomentosa Stib. PLoS ONE 2012, 7, e29678. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Tian, J.; Zhang, H.; Liao, H. Phytohormone regulation of root growth triggered by P deficiency or Al toxicity. J. Exp. Bot. 2016, 67, 3655–3664. [Google Scholar] [CrossRef] [PubMed]
- Kellermeier, F.; Amtmann, A. Analysis of the Root System Architecture of Arabidopsis Provides a Quantitative Readout of Crosstalk between Nutritional Signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yang, S.; Zhang, Y.; Liu, Y.; Meng, X.; Liang, Z. Metabolic profiles of three related Salvia species. Fitoterapia 2009, 80, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 Is Involved in Phosphate-Starvation Signaling and Excessive Phosphate Accumulation in Shoots of Plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Gao, W.Y.; Wang, J.; Cao, Y.; Zhao, Y.X.; Huang, L.Q.; Liu, C.X. Selection and optimization of a high-producing tissue culture of Panax ginseng C. A. Meyer. Acta Physiol. Plant. 2010, 32, 765–772. [Google Scholar] [CrossRef]
- Ding, K.; Pei, T.; Bai, Z.; Jia, Y.; Ma, P.; Liang, Z. SmMYB36, a Novel R2R3-MYB Transcription Factor, Enhances Tanshinone Accumulation and Decreases Phenolic Acid Content in Salvia miltiorrhiza Hairy Roots. Sci. Rep. 2017, 7, 5104. [Google Scholar] [CrossRef] [PubMed]
- Misson, J.; Raghothama, K.G.; Jain, A.; Jouhet, J.; Block, M.A.; Bligny, R.; Ortet, P.; Creff, A.; Somerville, S.; Rolland, N.; et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 2005, 102, 11934–11939. [Google Scholar] [CrossRef] [PubMed]
- Morcuende, R.; Bari, R.; Gibon, Y.; Zheng, W.; Pant, B.D.; Blã¤Sing, O.; Usadel, B.; Czechowski, T.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007, 30, 85–112. [Google Scholar] [CrossRef] [PubMed]
- Hassler, S.; Jung, B.; Lemke, L.; Novák, O.; Strnad, M.; Martinoia, E.; Neuhaus, H.E. Function of the Golgi-located phosphate transporter PHT4;6 is critical for senescence-associated processes in Arabidopsis. J. Exp. Bot. 2016, 67, 4671–4684. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive Genomic Identification and Expression Analysis of the Phosphate Transporter (PHT) Gene Family in Apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, C.; Ding, G.; Shi, L.; Xu, F. Genome-Wide Identification and Characterization of SPX Domain-Containing Members and Their Responses to Phosphate Deficiency in Brassica napus. Front. Plant Sci. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Huang, T.K.; Yang, S.Y.; Hong, Y.T.; Huang, S.M.; Wang, F.N.; Chiang, S.F.; Tsai, S.Y.; Lu, W.C.; Chiou, T.J. Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 2016, 7, 11095. [Google Scholar] [CrossRef] [PubMed]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; De, L.L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodríguez, J. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Han, H.; Zhang, K.; Wei, Z.; Yu, Y.; Wu, Z.; Ping, W. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 2014, 65, 859–870. [Google Scholar]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wei, J.; Wu, F.; Zhang, H.; Yang, D.; Liang, Z.; Jin, W. DsTRD: Danshen Transcriptional Resource Database. PLoS ONE 2016, 11, e0149747. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Gill, R.A.; Faisal, I.; Basharat, A.; Liu, H.; Xu, J.; He, S.; Zhou, W. Methyl Jasmonate Regulates Antioxidant Defense and Suppresses Arsenic Uptake in Brassica napus L. Front. Plant Sci. 2016, 7, 468. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Xing, B.; Yang, D.; Guo, W.; Liang, Z.; Yan, X.; Zhu, Y.; Liu, Y. Ag+ as a more effective elicitor for production of tanshinones than phenolic acids in Salvia miltiorrhiza hairy roots. Molecules 2015, 20, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Bao, Z.; Zhu, Z.; Ying, Q.; Qian, Q. Effects of Different Treatments of Salicylic Acid on Heat Tolerance, Chlorophyll Fluorescence, and Antioxidant Enzyme Activity in Seedlings of Cucumis sativa L. Plant Growth Regul. 2006, 48, 127–135. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Yang, D.; Xing, B.; Zhang, H.; Liang, Z. Salvia castanea Hairy Roots are More Tolerant to Phosphate Deficiency than Salvia miltiorrhiza Hairy Roots Based on the Secondary Metabolism and Antioxidant Defenses. Molecules 2018, 23, 1132. https://doi.org/10.3390/molecules23051132
Liu L, Yang D, Xing B, Zhang H, Liang Z. Salvia castanea Hairy Roots are More Tolerant to Phosphate Deficiency than Salvia miltiorrhiza Hairy Roots Based on the Secondary Metabolism and Antioxidant Defenses. Molecules. 2018; 23(5):1132. https://doi.org/10.3390/molecules23051132
Chicago/Turabian StyleLiu, Lin, Dongfeng Yang, Bingcong Xing, Haihua Zhang, and Zongsuo Liang. 2018. "Salvia castanea Hairy Roots are More Tolerant to Phosphate Deficiency than Salvia miltiorrhiza Hairy Roots Based on the Secondary Metabolism and Antioxidant Defenses" Molecules 23, no. 5: 1132. https://doi.org/10.3390/molecules23051132
APA StyleLiu, L., Yang, D., Xing, B., Zhang, H., & Liang, Z. (2018). Salvia castanea Hairy Roots are More Tolerant to Phosphate Deficiency than Salvia miltiorrhiza Hairy Roots Based on the Secondary Metabolism and Antioxidant Defenses. Molecules, 23(5), 1132. https://doi.org/10.3390/molecules23051132



