Carbazole Derivatives’ Binding to c-KIT G-Quadruplex DNA
Abstract
:1. Introduction
2. Results and Discussion
2.1. Ligands and Oligonucleotide
2.2. UV-Vis Absorption Spectroscopy
2.3. Fluorescence Spectroscopy
2.4. CD Spectroscopy
2.5. Binding of Ligands to c-KIT 1 G-Quadruplex
2.6. DNA Melting Studies
2.7. Comparison with c-MYC G-Quadruplex
2.8. Molecular Modeling Studies
3. Materials and Methods
3.1. Ligands
3.2. Oligonucleotide
3.3. Absorption Spectroscopy
3.4. Fluorescence Spectroscopy
3.5. Circular Dichroism
3.6. Ligand-Quadruplex Binding Study
3.7. Molecular Modeling Studies
3.8. Ligand Docking and Modeling
3.9. Molecular Dynamics Simulations
3.10. Interactions and Stability Analysis
3.11. Clustering
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Lacroix, L.; Douarre, C.; Temime-Smaali, N.; Trentesaux, C.; Riou, J.F.; Mergny, J.L. Targeting telomeres and telomerase. Biochimie 2008, 90, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.M.; Lu, Y.J.; Tan, J.H.; Huang, Z.S.; Wong, K.Y.; Gu, L.Q. G-quadruplexes: targets in anticancer drug design. Chem. Med. Chem. 2008, 3, 690–713. [Google Scholar] [CrossRef] [PubMed]
- Folini, M.; Gandellini, P.; Zaffaroni, N. Targeting the telosome: Therapeutic implications. Biochim. Biophys. Acta 2009, 1792, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Hurley, L.H.; Yang, D. Solution Structure of a 2:1 Quindolinec-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design. J. Am. Chem. Soc. 2011, 133, 17673–17680. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Propeller-Type Parallel-Stranded G-Quadruplexes in the Human c-myc Promoter. J. Am. Chem. Soc. 2004, 126, 8710–8716. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, T.; Pecinka, P.; Kubista, M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998, 26, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Lin, C.; Mathad, R.I.; Carver, M.; Yang, D. The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-nt Loop in K+ Solution. J. Am. Chem. Soc. 2014, 136, 1750–1753. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of Pseudosymmetrical G-Quadruplex and i-Motif Structures in the Proximal Promoter Region of the RET Oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Hatzakis, E.; Guo, K.; Carver, M.; Yang, D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: Insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013, 41, 10584–10592. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Lacroix, L.; Yue, D.J.E.; Lim, J.K.C.; Lim, J.M.W.; Phan, A.T. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. [Google Scholar] [CrossRef] [PubMed]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA Quadruplex Formation within the Human c-kit Oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Fernando, H.; Reszka, A.P.; Huppert, J.; Ladame, S.; Rankin, S.; Venkitaraman, A.R.; Neidle, S.; Balasubramanian, S. A Conserved Quadruplex Motif Located in a Transcription Activation Site of the Human c-kit Oncogene. Biochemistry 2006, 45, 7854–7860. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.K.; Haider, S.M.; Parkinson, G.N.; Neidle, S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007, 35, 5799–5808. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA Gquadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Balasubramanian, S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014, 25, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.T.D.; Varnai, P.; Bugaut, A.; Reszka, A.P.; Neidle, S.; Balasubramanian, S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009, 131, 13399–13409. [Google Scholar] [CrossRef] [PubMed]
- Kuryavyi, V.; Phan, A.T.; Patel, D.J. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010, 38, 6757–6773. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012, 40, 4691–4700. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Kuang, W.J.; Yang, F.T.; Coussens, L.; Munemitsu, S.; Dull, T.J.; Chen, E.; Schlessinger, J.; Francke, U.; Ullrich, A. Human proto-oncogene c-kit: A new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987, 6, 3341–3351. [Google Scholar] [PubMed]
- Roskoski, R.J. Structure and regulation of Kit protein-tyrosine kinase, the stem cell factor receptor. Biochem. Biophys. Res. Commun. 2005, 37, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Edling, C.E.; Hallberg, B. c-Kit—A hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol. 2007, 39, 1995–1998. [Google Scholar] [CrossRef] [PubMed]
- Gregory, B.E.; Bartlett, E.; Kiupel, M.; Hayes, S.; Yuzbasiyan, G.V. Canine and human gastrointestinal stromal tumors display similar mutations in c-KIT exon 11. BMC Cancer 2010, 10, 1–9. [Google Scholar]
- Lennartsson, J.; Rönnstrand, L. Stem cell factor receptor/c-kit: from basic science to clinical implications. Physiol Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Zhou, G.B.; Yin, T.; Chen, B.; Shi, J.Y.; Liang, W.X.; Jin, X.L.; You, J.H.; Yang, G.; Shen, Z.X.; et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: Implication in stepwise leukemogenesis and response to Gleevec. J. Proc. Natl. Acad. Sci. USA 2005, 102, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Grande, A.; Zucchini, P.; Manfredini, R.; Tagliafico, E.; Rossi, E.; Temperani, P.; Torelli, G.; Emilia, G.; Torelli, U. Overexpression of c-kit in a leukemic cell population carrying a trisomy 4 and its relationship with the proliferative capacity. Leuk. Lymphoma 1993, 9, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Lasota, J. KIT (CD117): A review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl. Immunohistochem. 2005, 13, 205–220. [Google Scholar] [CrossRef]
- Simak, R.; Capodieci, P.; Cohen, D.W.; Fair, W.R.; Scher, H.; Melamed, J.; Drobnjak, M.; Heston, W.D.; Stix, U.; Steiner, G.; et al. Expression of c-kit and kit-ligand in benign and malignant prostatic tissues. Histol. Histopathol. 2000, 15, 365–374. [Google Scholar] [PubMed]
- Micke, P.; Hengstler, J.G.; Albrecht, H.; Faldum, A.; Bittinger, F.; Becker, K.; Wiewrodt, R.; Fischer, B.; Buhl, R. c-kit Expression in Adenocarcinomas of the Lung. Tumour Biol. 2004, 25, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, A.; Sawai, H.; Takahashi, H.; Ochi, N.; Matsuo, Y.; Funahashi, H.; Sato, M.; Okada, Y.; Takeyama, H.; Manabe, T. The stem cell factor/c-kit receptor pathway enhances proliferation and invasion of pancreatic cancer cells. Mol. Cancer 2006, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, A.; Summersgill, B.; Grygalewicz, B.; Gillis, A.J.; Stoop, J.; van Gurp, R.J.; Dennis, N.; Fisher, C.; Huddart, R.; Cooper, C.; et al. Amplification and overexpression of the KIT gene Is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults. Cancer Res. 2005, 65, 8085–8089. [Google Scholar] [CrossRef] [PubMed]
- Smalley, K.S.M.; Nathanson, K.L.; Flaherty, K.T. Genetic subgrouping of melanoma reveals new opportunities for targeted therapy. Cancer Sci. 2009, 69, 3241–3244. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, S.; Fukasawa, T.; Chong, J.M.; Tanaka, A.; Fukayama, M. C-kit gene abnormalities in gastrointestinal stromal tumors (Tumors of Interstitial Cells of Cajal). Cancer Sci. 1999, 90, 1321–1328. [Google Scholar]
- Sattler, M.; Salgia, R. Targeting c-Kit mutations: basic science to novel therapies. Leukaemia Res. 2004, 28, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Tuveson, D.A.; Willis, N.A.; Jacks, T.; Griffin, T.D.; Singer, S.; Fletcher, C.D.M.; Fletcher, J.A.; Demetri, G.D. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene 2001, 20, 5054–5058. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.C.; Corless, C.L.; Blanke, C.D.; Demetri, G.D.; Joensuu, H.; Roberts, P.J.; Eisenberg, B.L.; von Mehren, M.; Fletcher, C.D.M.; Sandau, K.; et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J. Clin. Oncol. 2006, 24, 4764–4774. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, S.A. Acquired resistance to drugs targeting receptor tyrosine kinases. Biochem. Pharmacol. 2012, 82, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Bejugam, M.; Sewitz, S.; Shirude, P.S.; Rodriguez, R.; Shahid, R.; Balasubramanian, S. Trisubstituted isoalloxazines as a new class of G-Quadruplex binding ligands: Small molecule regulation of c-kit oncogene expression. J. Am. Chem. Soc. 2007, 129, 12926–12927. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.W.; Liu, M.-C.; Lee, K.-D.; Chang, K.W.; Yang, Y.-T.; Tung, H.-W.; Fox, K.R. Synthesis of bisquinoline–pyrrole oligoamide as G-quadruplex binding ligand. Tetrahedron 2012, 68, 5453–5457. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, C.-X.; Yan, J.-W.; Hou, J.-Q.; Chen, S.-B.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S.; Tan, J.-H. Synthesis and evaluation of quinazolone derivatives as a new class of c-KIT G-Quadruplex binding ligands. ACS Med. Chem. Lett. 2013, 4, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Garner, T.P.; Williams, H.E.L.; Gluszyk, K.I.; Roe, S.; Oldham, N.J.; Stevens, M.F.G.; Moses, J.E.; Searle, M.S. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org. Biomol. Chem. 2009, 7, 4194–4200. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.C.; Larsen, A.F.; Abdikadir, F.H.; Ulven, T. Phenanthroline-2,9-bistriazoles as selective G-quadruplex ligands. Eur. J. Med. Chem. 2014, 72, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Gunaratnam, M.; Swank, S.; Haider, S.M.; Galesa, K.; Reszka, A.P.; Beltran, M.; Cuenca, F.; Fletcher, J.A.; Neidle, S. Targeting human gastrointestinal stromal tumor cells with a Quadruplex-Binding small molecule. J. Med. Chem. 2009, 52, 3774–3783. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lin, S.; Yuan, G. Spectroscopic probing of recognition of the G-quadruplex in c-kit promoter by small-molecule natural products. Int. J. Biol. Macromolec. 2012, 50, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.-H.; Jin, J.; Li, J.; Wang, Y.; Zhu, S.-H.; Lu, Y.-J.; Ou, T.-M.; Huang, Z.-S.; Huang, M.; Huang, Z.-Y. The G-quadruplex ligand, SYUIQ-FM05, targets proto-oncogene c-kit transcription and induces apoptosis in K562 cells. Pharm. Biol. 2013, 51, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-F.; Qin, Q.-P.; Qin, J.-L.; Liu, Y.-C.; Huang, K.-B.; Li, Y.-L.; Meng, T.; Zhang, G.-H.; Peng, Y.; Luo, X.-J.; et al. Stabilization of G-Quadruplex DNA, inhibition of telomerase activity, and tumor cell apoptosis by organoplatinum (II) complexes with oxoisoaporphine. J. Med. Chem. 2015, 58, 2159–2179. [Google Scholar] [CrossRef] [PubMed]
- Jantos, K.; Rodriguez, R.; Ladame, S.; Shirude, P.S.; Balasubramanian, S. Oxazole-based peptide macrocycles: A new class of G-Quadruplex binding ligands. J. Am. Chem. Soc. 2006, 128, 13662–13663. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.M.; Reszka, A.P.; Gunaratnam, M.; Haider, S.M.; Howard, P.W.; Fox, K.R.; Neidle, S.; Thurston, D.E. Biaryl polyamides as a new class of DNA quadruplex-binding ligands. Chem. Commun. 2009, 4097–4099. [Google Scholar] [CrossRef] [PubMed]
- Bejugam, M.; Gunaratnam, M.; Müller, S.; Sanders, D.A.; Sewitz, S.; Fletcher, J.A.; Neidle, S.; Balasubramanian, S. Targeting the c-Kit promoter G-quadruplexes with 6-substituted indenoisoquinolines. ACS Med. Chem. Lett. 2010, 1, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Diveshkumar, K.V.; Sakrikar, S.; Harikrishna, S.; Dhamodharan, V.; Pradeepkumar, P.I. Targeting promoter G-Quadruplex DNAs by indenopyrimidine-Based ligands. Chem. Med. Chem. 2014, 9, 2754–2765. [Google Scholar] [CrossRef] [PubMed]
- McLuckie, K.I.E.; Waller, Z.A.E.; Sanders, D.A.; Alves, D.; Rodriguez, R.; Dash, J.; McKenzie, G.J.; Venkitaraman, A.R.; Balasubramanian, S. G-Quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J. Am. Chem. Soc. 2011, 133, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Diveshkumar, K.V.; Sakrikar, S.; Rosu, F.; Harikrishna, S.; Gabelica, V.; Pradeepkumar, P.I. Specific stabilization of c-MYC and c-KIT G-Quadruplex DNA structures by indolylmethyleneindanone scaffolds. Biochemistry 2016, 55, 3571–3585. [Google Scholar] [CrossRef] [PubMed]
- Dash, J.; Nath Das, R.; Hegde, N.; Dan Pantos, G.; Shirude, P.S.; Balasubramanian, S. Synthesis of bis-indole carboxamides as G-Quadruplex stabilizing and inducing ligands. Chem. Eur. J. 2012, 18, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, V.; Harikrishna, S.; Bhasikuttan, A.C.; Pradeepkumar, P.I. Topology specific stabilization of promoter over telomeric G-Quadruplex DNAs by bisbenzimidazole carboxamide derivatives. ACS Chem. Biol. 2015, 10, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Ren, L.; Gao, N. Interactions of terpyridines and their Pt(II) complexes with G-quadruplex DNAs and telomerase inhibition. Int. J. Biol. Macromol. 2013, 57, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Waller, Z.A.E.; Sewitz, S.A.; Hsu, S.-T.D.; Balasubramanian, S. A small molecule that disrupts G-Quadruplex DNA structure and enhances gene expression. J. Am. Chem. Soc. 2009, 131, 12628–12633. [Google Scholar] [CrossRef] [PubMed]
- Manaye, S.; Eritja, R.; Aviñó, A.; Jaumot, J.; Gargallo, R. Porphyrin binding mechanism is altered by protonation at the loops in G-quadruplex DNA formed near the transcriptional activation site of the human c-kit gene. Biochim. Biophys. Acta 2012, 1820, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Petenzi, M.; Verga, D.; Largy, E.; Hamon, F.; Doria, F.; Teulade-Fichou, M.-P.; Guédin, A.; Mergny, J.-L.; Mella, M.; Freccero, M. Cationic pentaheteroaryls as selective G-Quadruplex ligands by solvent-free microwave-assisted synthesis. Chem. Eur. J. 2012, 18, 14487–14496. [Google Scholar] [CrossRef] [PubMed]
- Alzeer, J.; Luedtke, N.W. PH-Mediated fluorescence and G-Quadruplex binding of amido phthalocyanines. Biochemistry 2010, 49, 4339–4348. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.-W.; Zhang, D.; Zhang, L.-X.; Hao, Y.-H.; Zhou, X.; Tan, Z. Dissecting the strand folding orientation and formation of G-Quadruplexes in single- and double-stranded nucleic acids by ligand-induced photocleavage footprinting. J. Am. Chem. Soc. 2011, 133, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, C.; Minarini, A.; Tumiatti, V.; Moraca, F.; Parrotta, L.; Alcaro, S.; Rigo, R.; Sissi, C.; Gunaratnam, M.; Ohnmacht, S.A.; et al. Macrocyclic naphthalene diimides as G-quadruplex binders. Bioorg. Med. Chem. 2015, 23, 3819–3830. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M; Fujii, S.; Sato, S.; Okauchi, T.; Takenaka, S. A selective G-Quadruplex DNA-stabilizing ligand based on a cyclic naphthalene diimide derivative. Molecules 2015, 20, 10963–10979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorzan, E.; Da Ros, S.; Musetti, C.; Shahidian, L.Z.; Coelho, N.F.R.; Bonsembiante, F.; Létard, S.; Gelain, M.E.; Palumbo, M.; Dubreuil, P.; et al. Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncoterget 2016, 7, 21658–21675. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A. Biological potential of carbazole derivatives. Eur. J. Med. Chem. 2015, 94, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole ligands as c-myc G-quadruplex binders. Int. J. Biol. Macromol. 2018, 114, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Bajor, K.; Czerwińska, I.; Kalet, D.; Juskowiak, B. The synthesis and spectral properties of new DNA binding ligands. Tetrahedron Lett. 2010, 51, 5415–5418. [Google Scholar] [CrossRef]
- Głuszyńska, A.; Rajczak, E.; Juskowiak, B. The synthesis and spectral properties of new carbazole ligand, potential DNA intercalator. In Proceedings of the 40th International Conference of Slovak Society of Chemical Engineering, Tatranské Matliare, Slovakia, 31 May 2013; pp. 1349–1354. [Google Scholar]
- Głuszyńska, A.; Rajczak, E.; Juskowiak, B. Synthesis and spectroscopic characterisation of (E)-2-(2-(9-(4-(1H-1,2,4-triazol-1-yl)butyl)-9H-carbazol-3-yl)vinyl)-3-ethylbenzo[d]thiazol-3-ium, a new ligand and potential DNA intercalator. Chem. Pap. 2013, 67, 1231–1239. [Google Scholar] [CrossRef]
- Głuszyńska, A.; Rajczak, E.; Kosman, J.; Juskowiak, B. Interactions of New Carbazole Ligands, Potential Inhibitors of Telomerase, with Different Structures of DNA: A Comparative Study. In Proceedings of the 40th International Conference of Slovak Society of Chemical Engineering, Tatranské Matliare, Slovakia, 27–31 May 2013; pp. 1326–1332. [Google Scholar]
- Głuszyńska, A.; Burzyńska, K.; Rajczak, E.; Juskowiak, B. Tatranské Matliare, Slovakia. Interaction of N-phenyl carbazole Ligands with Double Stranded structures of DNA. In Proceedings of the 40th International Conference of Slovak Society of Chemical Engineering, Tatranské Matliare, Slovakia, 31 May 2013; pp. 526–530. [Google Scholar]
- Saengkhae, C.; Salerno, M.; Adès, D.; Siove, A.; Le Moyec, L.; Migonney, V.; Garnier-Suillerot, A. Ability of carbazole salts, inhibitors of Alzheimer β-amyloid fibril formation, to cross cellular membranes. Eur. J. Pharmacol. 2007, 559, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Kowal, A.; Kolasa, A.; Rajczak, E.; Juskowiak, B.; Rubiś, B.; Takenaka, S. Synthesis and characterization of fluorescent DNA binding carbazole ligand. Manuscript in preparation.
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Quadruplex Nucleic Acids, 1st ed.; Neidle, S., Balasubramanian, S., Eds.; Publisher: Royal Society of Chemistry, London, UK, 2006; Volume I, pp. 81–99. [Google Scholar]
- Bhattacharjee, A.J.; Ahluwalia, K.; Taylor, S.; Jin, O.; Nicoludis, J.M.; Buscaglia, R.; Chaires, J.B.; Kornfilt, D.J.P.; Marquardt, D.G.S.; Yatsunyk, L.A. Induction of G-quadruplex DNA structure by Zn (II) 5,10,15,20-tetrakis (N-methyl-4-pyridyl)porphyrin. Biochimie 2011, 93, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Uno, T.; Ishikawa, Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005, 13, 2423–2430. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Liu, J.; Li, J.; Zheng, K.-C.; Huang, X.-M.; Tan, C.-P.; Chen, L.-M.; Ji, L.-N. Synthesis, characterization and DNA-binding of novel chiral complexes D- and K-[Ru(bpy)2L] 2+ (L = o-mopip and p-mopip). J. Inorg. Biochem. 2006, 100, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; An, Y.; Zhang, L.; Chen, H.-Y.; Han, Y.; Wang, Y.-J.; Mao, Z.-W.; Ji, L.-N. Studies on synthesis, characterization, and G-quadruplex binding of Ru (II) complexes containing two dppz ligands. J. Inorg. Biochem. 2011, 105, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Kumar, K.; Kaulage, M.; Muniyappa, K.; Bhattacharya, S. Design and synthesis of new benzimidazole−carbazole conjugates for the stabilization of human telomeric DNA, telomerase inhibition, and their selective action on cancer cells. J. Med. Chem. 2014, 57, 6973–6988. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Kumar, K.; Muniyappa, K.; Bhattacharya, S. New dimeric carbazole–benzimidazole mixed ligands for the stabilization of human telomeric G-quadruplex DNA and as telomerase inhibitors. A remarkable influence of the spacer. Org. Biomol. Chem. 2015, 13, 8335–8348. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Trotta, R.; Pieraccini, S.; De Tito, S.; Perone, R.; Randazzo, A.; Spada, G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem., 2010, 8, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.-H.; Bochkareva, E.; Bochkarev, A.; Gray, D.M. Circular dichroism spectra and electrophoretic mobility shift assays show that human replication protein a binds and melts intramolecular G-Quadruplex structures. Biochemistry 2009, 48, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Crosby, G.A.; Demas, J.N. Measurement of photoluminescence quantum yields. Review. J. Phys. Chem. 1971, 75, 991–1024. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Phan, A.-T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Fox, K.R. Quadruplex melting. Methods 2007, 43, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Marchán, I.; Svozil, D.; Sponer, J.; Cheatham, T.E.; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [PubMed]
- Zgarbová, M.; Otyepka, M.; Šponer, J.; Mládek, A.; Banáš, P.; Cheatham, T.E.; Jurečka, P. Refinement of the cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef] [PubMed]
- Banáš, P.; Hollas, D.; Zgarbová, M.; Jurečka, P.; Orozco, M.; Cheatham, T.E.; Šponer, J.; Otyepka, M. Performance of molecular mechanics force fields for RNA simulations: stability of UUCG and GNRA hairpins. J. Chem. Theory Comput. 2010, 6, 3836–3849. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Abagyan, R.; Totrov, M.; Kuznetsov, D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994, 15, 488–506. [Google Scholar] [CrossRef]
- Harvey, M.J.; Giupponi, G.; Fabritiis, G.D. ACEMD: Accelerating biomolecular dynamics in the microsecond time Scale. J. Chem. Theory Comput. 2009, 5, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. Gromacs: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Size-independent comparison of protein three-dimensional structures. Proteins 2004, 22, 273–283. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


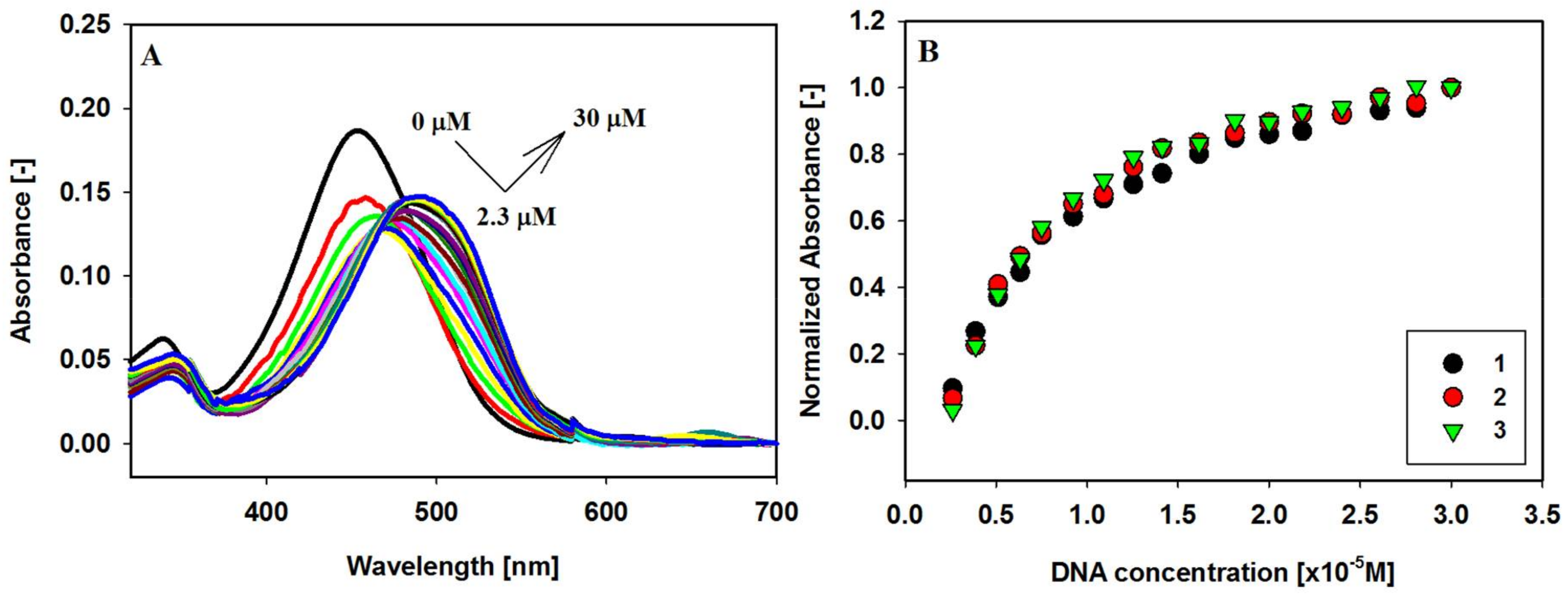


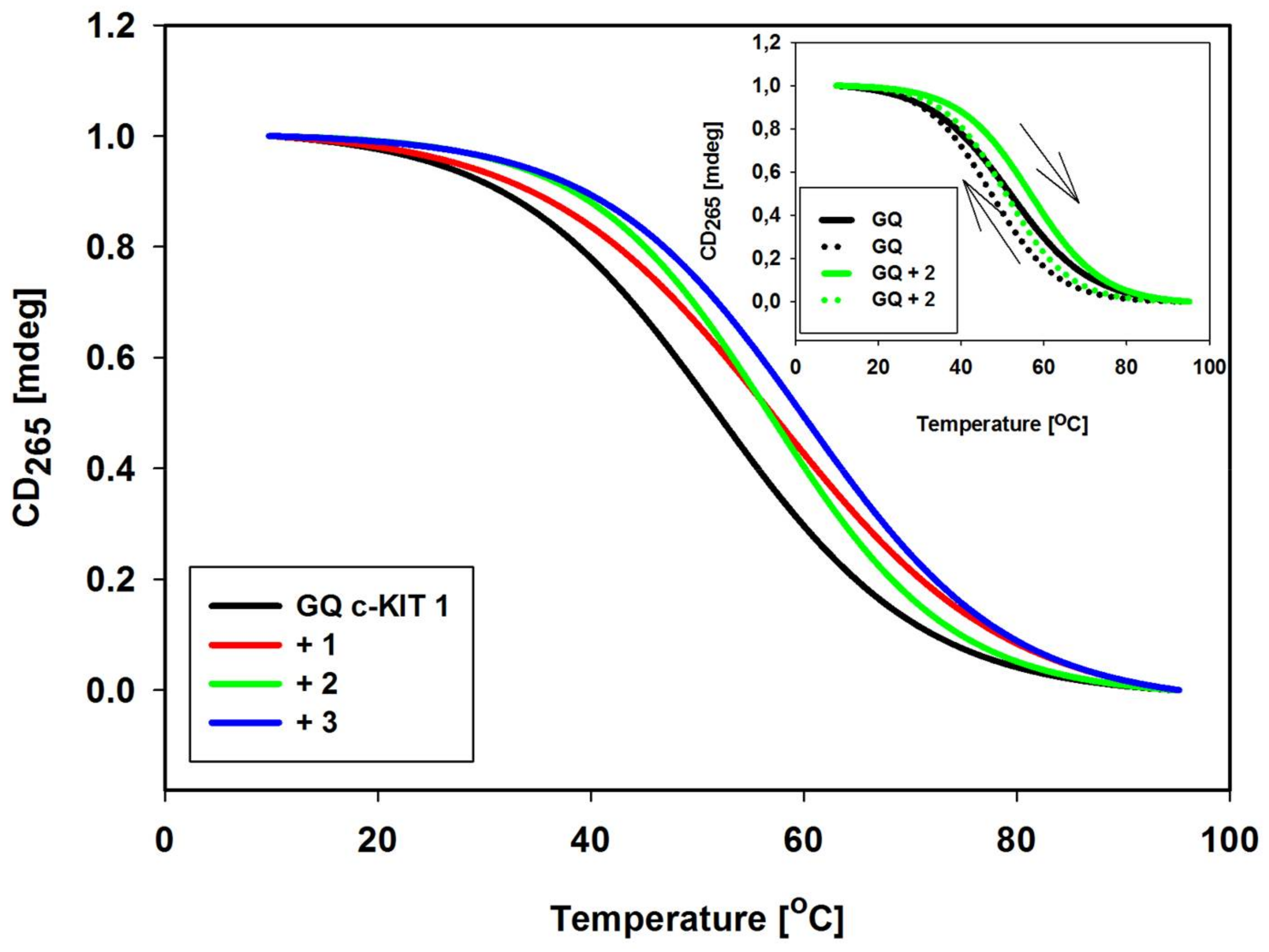
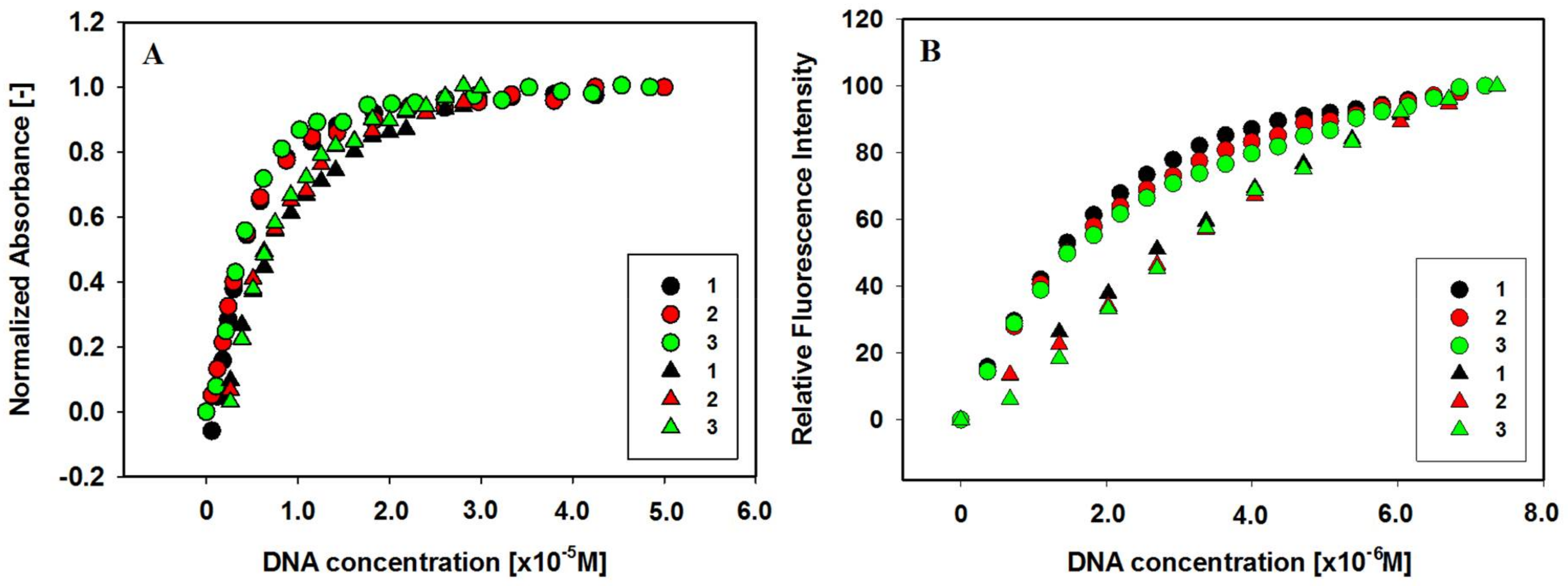


| Ligands | Δλmax (nm) a | Hypochromicity (%) b | Hyperchromicity (%) b |
|---|---|---|---|
| 1 | 36 | 35 | 15 |
| 2 | 38 | 30 | 16 |
| 3 | 42 | 31 | 25 |
| Ligands | Benesi-Hildebrand Method, nKb (× 105 M−1) | |
|---|---|---|
| Spectrophotometric Titration | Fluorescence Titration | |
| 1 | 1.1 ± 0.1 | 1.5 ± 0.2 |
| 2 | 1.4 ± 0.1 | 1.2 ± 0.1 |
| 3 | 2.0 ± 0.1 | 1.4 ± 0.1 |
| Derivative | Tm (°C) a | ΔTm (°C) b | Tm (°C) c | ΔTm (°C) b |
|---|---|---|---|---|
| No drug | 51.8 | - | 77.9 | - |
| 1 | 57.7 | 5.9 | 79.9 | 2.0 |
| 2 | 57.2 | 5.4 | 79.9 | 2.0 |
| 3 | 60.2 | 8.4 | 82.2 | 4.3 |
| Effect | Ligands | c-MYC a | c-KIT |
|---|---|---|---|
| Δλmax (nm) b | 1 | 57 | 36 |
| 2 | 52 | 38 | |
| 3 | 52 | 42 | |
| Hypochromicity (%) c | 1 | 30 | 35 |
| 2 | 24 | 30 | |
| 3 | 28 | 31 | |
| Hyperchromicity (%) c | 1 | 32 | 15 |
| 2 | 14 | 16 | |
| 3 | 18 | 25 | |
| Fluorescence titration | 1 | 6.9 ± 0.1 | 1.5 ± 0.2 |
| 2 | 4.0 ± 0.1 | 1.2 ± 0.1 | |
| 3 | 3.9 ± 0.1 | 1.4 ± 0.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole Derivatives’ Binding to c-KIT G-Quadruplex DNA. Molecules 2018, 23, 1134. https://doi.org/10.3390/molecules23051134
Głuszyńska A, Juskowiak B, Kuta-Siejkowska M, Hoffmann M, Haider S. Carbazole Derivatives’ Binding to c-KIT G-Quadruplex DNA. Molecules. 2018; 23(5):1134. https://doi.org/10.3390/molecules23051134
Chicago/Turabian StyleGłuszyńska, Agata, Bernard Juskowiak, Martyna Kuta-Siejkowska, Marcin Hoffmann, and Shozeb Haider. 2018. "Carbazole Derivatives’ Binding to c-KIT G-Quadruplex DNA" Molecules 23, no. 5: 1134. https://doi.org/10.3390/molecules23051134






