Design, Synthesis and Biological Evaluation of Novel Phenylsulfonylurea Derivatives as PI3K/mTOR Dual Inhibitors
Abstract
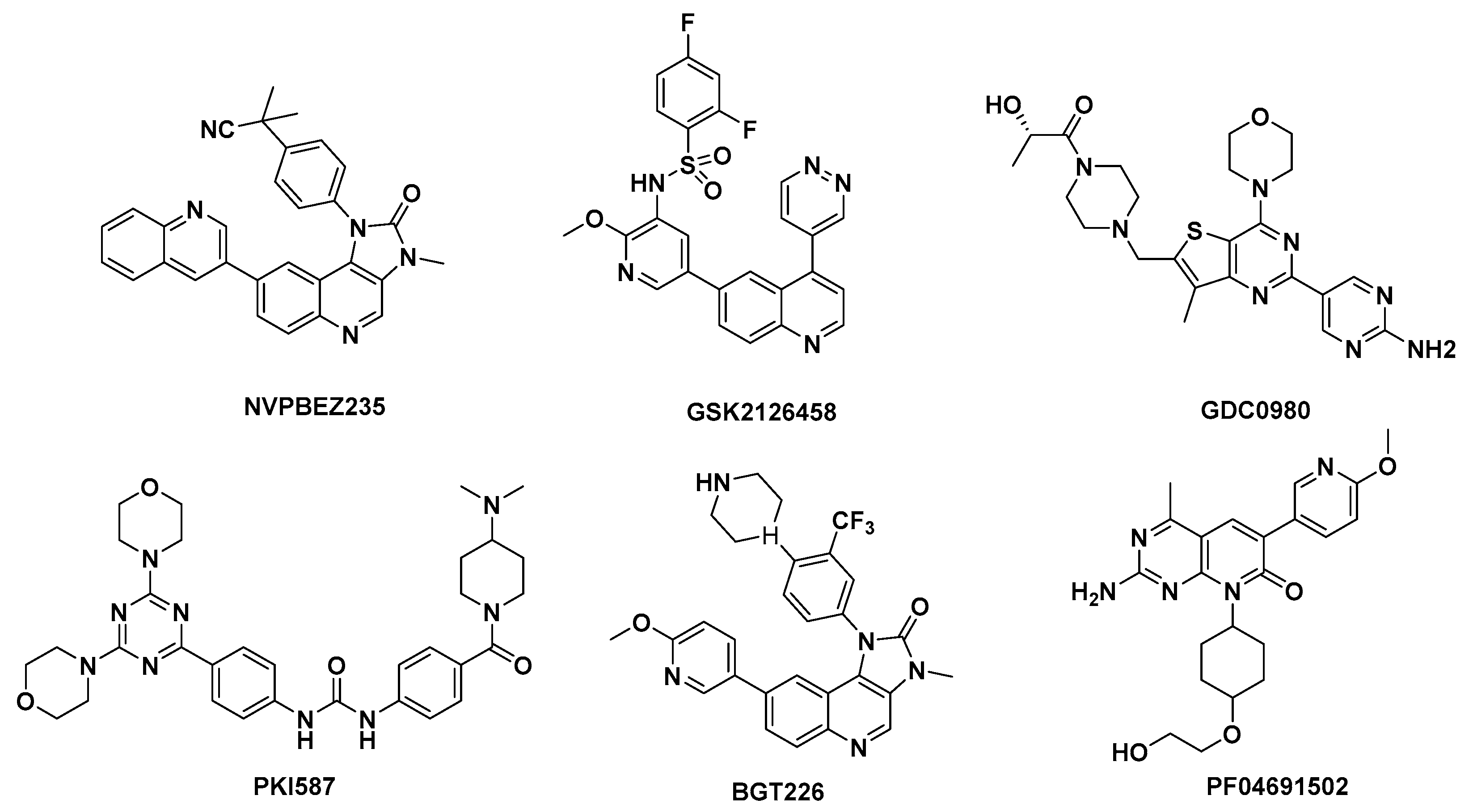
:1. Introduction
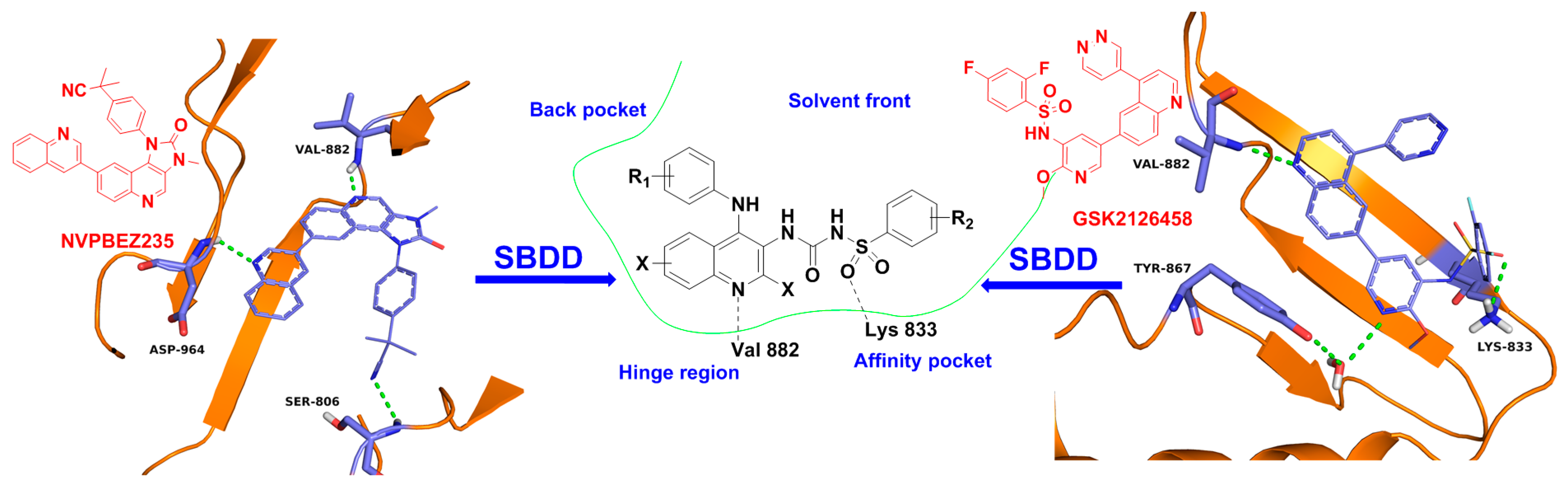
2. Results and Discussion
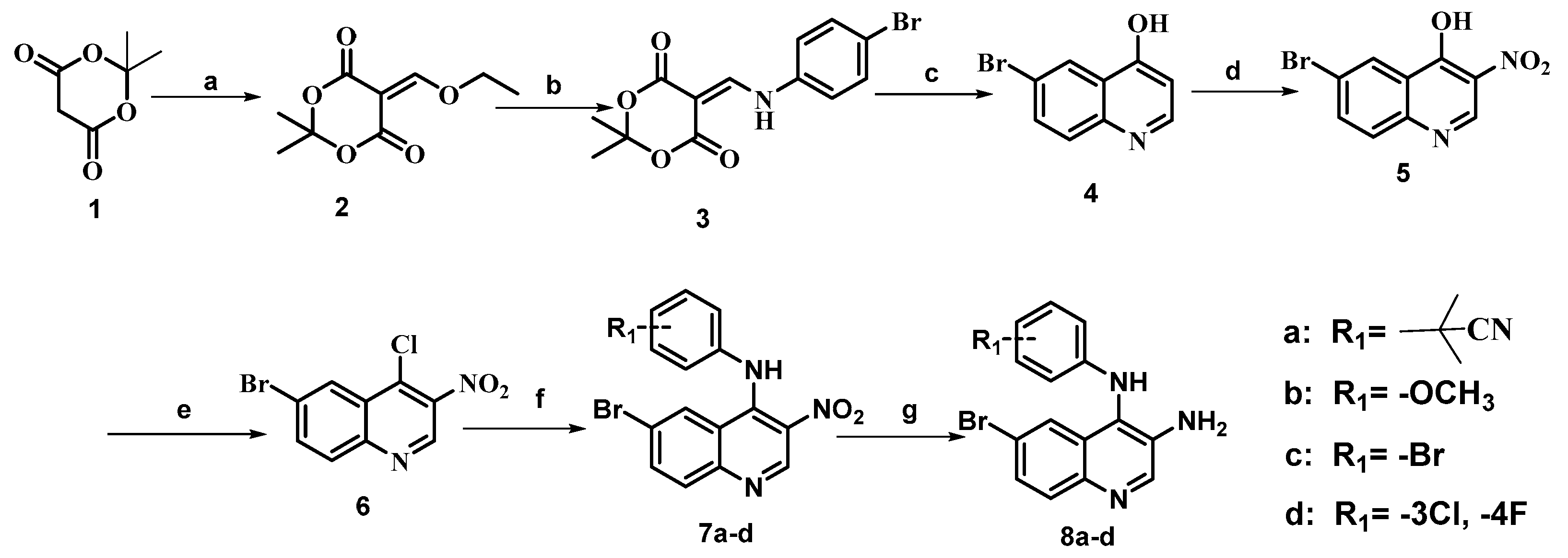
2.1. Chemistry
2.2. Biological Discussion
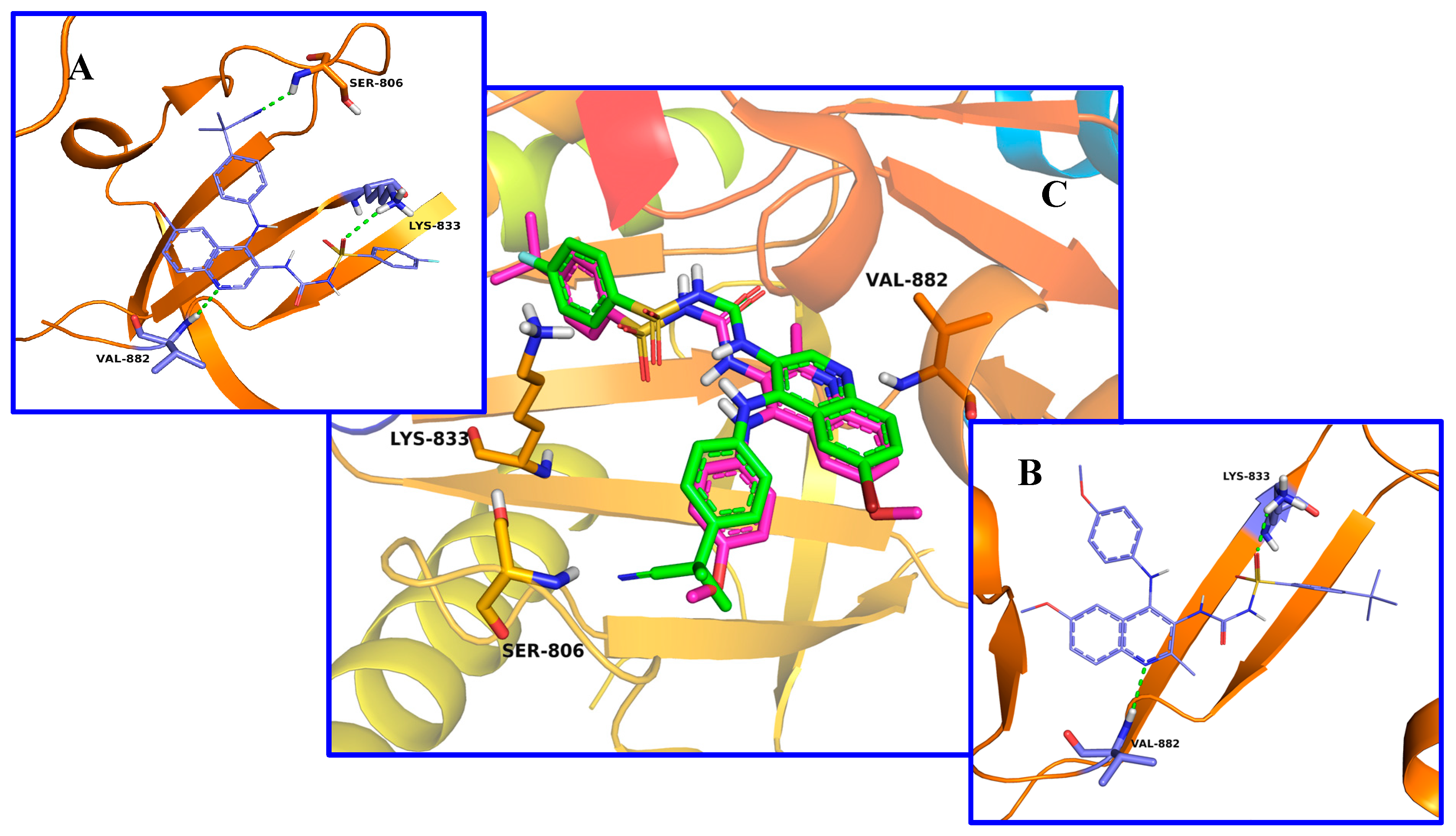
2.3. Molecular Docking Study
3. Experimental Section
3.1. General Information
3.2. Chemistry
3.2.1. General Procedure for the Preparation of Compounds 8a–d, 14a–d and 18a–d
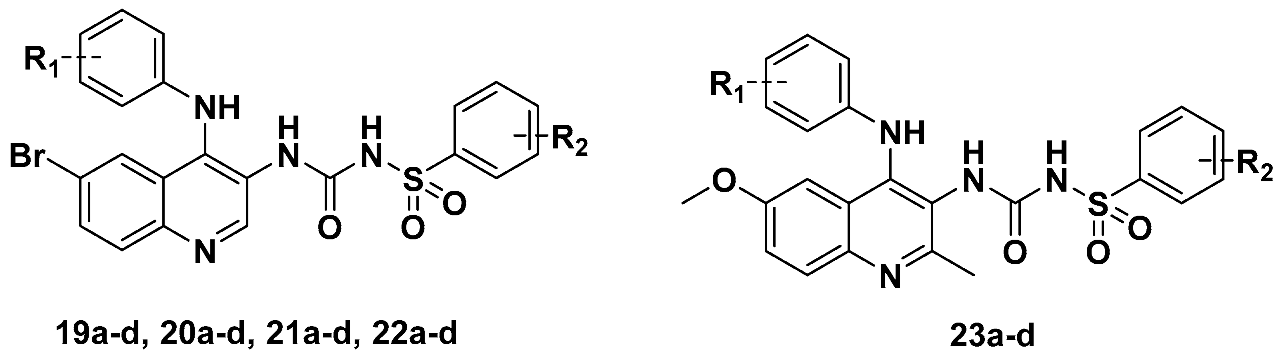
3.2.2. General Procedure for Preparation of Target Compounds 19a–d, 20a–d, 21a–d, 22a–d and 23a–d
3.3. PI3Kα Kinase Assay
3.4. mTOR Kinase Assay
3.5. Cytotoxicity Assay In Vitro
3.6. Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, B.; Sun, A.; Jiang, W.; Thrasher, J.B.; Terranova, P. PI-3 kinase p110beat: A therapeutic target in advanced prostate cancers. Am. J. Clin. Exp. Urol. 2014, 2, 188–198. [Google Scholar] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leevers, S.J.; Vanhaesebroeck, B.; Waterfield, M.D. Signalling through phosphoinositide 3-kinases: The lipids take centre stage. Curr. Opin. Cell Biol. 1999, 11, 219–225. [Google Scholar] [CrossRef]
- Giordanetto, F.; Wallberg, A.; Ghosal, S.; Iliefski, T.; Cassel, J.; Yuan, Z.Q.; Wachenfeldt, v.H.; Andersen, S.M.; Inghardt, T.; Tunek, A. Discovery of phosphoinositide 3-kinases (PI3K) p110β isoform inhibitor 4-[2-hydroxyethyl(1-naphthylmethyl)amino]-6-[(2S)-2-methylmorpholin-4-yl]-1H-pyrimidin-2-one, an effective antithrombotic agent without associated bleeding and insulin resistance. Bioorg. Med. Chem. Lett. 2012, 22, 6671–6676. [Google Scholar] [CrossRef] [PubMed]
- Certal, V.; Halley, F.; Vironeoddos, A.; Delorme, C.; Karlsson, A.; Rak, A.; Thompson, F.; Filocherommé, B.; Elahmad, Y.; Carry, J.C. Discovery and optimization of new benzimidazole- and benzoxazole-pyrimidone selective PI3Kβ inhibitors for the treatment of phosphatase and TENsin homologue (PTEN)-deficient cancers. J. Med. Chem. 2012, 55, 4788–4805. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, B.T.; Smith, D.L.; Ram, P.T.; Lu, Y.; Mills, G.B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005, 4, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Ma, X.; Hu, Y. Furthering the design and the discovery of small molecule ATP-competitive mTOR inhibitors as an effective cancer treatment. Expert Opin. Drug Dis. 2013, 8, 991–1012. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Tu, Z.C.; Long, Z.J.; Liu, Q.; Lu, G. Discovery of 2-(2-aminopyrimidin-5-yl)-4-morpholino-N-(pyridin-3-yl)quinazolin-7-amines as novel PI3K/mTOR inhibitors and anticancer agents. Eur. J. Med. Chem. 2015, 108, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Mader, M.M.; Cook, J.A.; Iversen, P.; Ajamie, R.; Perkins, E.; Bloem, L.; Yip, Y.Y.; Barda, D.A.; Waid, P.P. Characterization of LY3023414, a novel PI3K/mTOR dual inhibitor eliciting transient target modulation to impede tumor growth. Mol. Cancer Ther. 2016, 15, 2344–2356. [Google Scholar] [CrossRef] [PubMed]
- Asati, V.; Mahapatra, D.K.; Bharti, S.K. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: Structural and pharmacological perspectives. Eur. J. Med. Chem. 2016, 109, 314–341. [Google Scholar] [CrossRef] [PubMed]
- Serra, V.; Markman, B.M.; Eichhorn, P.; Valero, V.; Guzman, M.; Botero, M.; Llonch, E.; Atzori, F.; Di-Cosimo, S.; Maira, M. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008, 68, 8022–8030. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.D.; Adams, N.D.; Burgess, J.L.; Chaudhari, A.M.; Darcy, M.G.; Donatelli, C.A.; Luengo, J.I.; Newlander, K.A.; Parrish, C.A.; Ridgers, L.H. Discovery of GSK2126458, a highly potent inhibitor of PI3K and the mammalian target of rapamycin. ACS Med. Chem. Lett. 2010, 1, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Sutherlin, D.P.; Bao, L.; Berry, M.; Castanedo, G.; Chuckowree, I.; Dotson, J.; Folks, A.; Friedman, L.; Goldsmith, R.; Gunzner, J. Discovery of a potent, selective, and orally available class I phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) kinase inhibitor (GDC-0980) for the treatment of cancer. J. Med. Chem. 2011, 54, 7579–7587. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, A.M.; Dehnhardt, C.M.; Delos, S.E.; Chen, Z.; Dos, S.O.; Ayralkaloustian, S.; Khafizova, G.; Brooijmans, N.; Mallon, R.; Hollander, I. Bis(morpholino-1,3,5-triazine) derivatives: Potent adenosine 5′-triphosphate competitive phosphatidylinositol-3-kinase/mammalian target of rapamycin inhibitors: Discovery of compound 26 (PKI-587), a highly efficacious dual inhibitor. J. Med. Chem. 2010, 53, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.Y.; Tsai, S.Y.; Wu, C.M.; Yen, C.J.; Chuang, B.F.; Chang, J.Y. Novel phosphoinositide 3-kinase/mTOR dual inhibitor, NVP-BGT226, displays potent growth-inhibitory activity against human head and neck cancer cells in vitro and in vivo. Clin. Cancer Res. 2011, 17, 7116–7126. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Bagrodia, S.; Bailey, S.; Edwards, M.; Hoffman, J.; Hu, Q.; Kania, R.; Knighton, D.R.; Marx, M.A.; Ninkovic, S. Discovery of the highly potent PI3K/mTOR dual inhibitor PF-04691502 through structure based drug design. Med. Chem. Commun. 2010, 1, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Xi, L.; Zhang, J.Q.; Liu, Z.C.; Zhang, J.H.; Yan, J.F.; Jin, Y.; Lin, J. Novel 5-anilinoquinazoline-8-nitro derivatives as inhibitors of VEGFR-2 tyrosine kinase: Synthesis, biological evaluation and molecular docking. Org. Biomol. Chem. 2013, 11, 4367–4378. [Google Scholar] [CrossRef] [PubMed]
- Hoegenauer, K.; Soldermann, N.; Stauffer, F.; Furet, P.; Graveleau, N.; Smith, A.B.; Hebach, C.; Hollingworth, G.J.; Lewis, I.; Gutmann, S. Discovery and pharmacological characterization of novel quinazoline-based PI3K delta-selective inhibitors. ACS Med. Chem. Lett. 2016, 7, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.R.; Guru, S.K.; Joshi, P.; Mahajan, G.; Mintoo, M.J.; Kumar, V.; Bharate, S.S.; Mondhe, D.M.; Vishwakarma, R.A.; Bhushan, S. 6-Aryl substituted 4-(4-cyanomethyl) phenylamino quinazolines as a new class of isoform-selective PI3K-alpha inhibitors. Eur. J. Med. Chem. 2016, 122, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Hei, Y.Y.; Zhang, H.; Shen, Y.; Zhang, S.Q. Design and synthesis of novel 6-aryl substituted 4-anilinequinazoline derivatives as potential PI3Kδ inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 1972–1977. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 19a–d, 20a–d, 21a–d, 22a–d and 23a–d are available from the authors. |


| Compound | R1 | R2 | IC50 (µM) a | |||
|---|---|---|---|---|---|---|
| HepG-2 | A549 | PC-3 | MCF-7 | |||
| 19a | pivalonitrile | -F | 33.28 ± 1.52 | 38.52 ± 1.04 | 66.62 ± 1.28 | 3.88 ± 0.58 |
| 19b | pivalonitrile | -Cl | 30.22 ± 1.45 | >100 | 34.31 ± 1.32 | 15.10 ± 1.1 |
| 19c | pivalonitrile | -CH3 | 31.25 ± 1.50 | 28.96 ± 1.31 | 40.59 ± 1.11 | 3.96 ± 0.59 |
| 19d | pivalonitrile | Tert-Butyl | >100 | 30.58 ± 1.45 | 68.25 ± 1.32 | 9.95 ± 0.99 |
| 20a | -OCH3 | -F | 20.34 ± 1.04 | 26.34 ± 1.13 | >100 | 7.65 ± 0.88 |
| 20b | -OCH3 | -Cl | 22.56 ± 1.12 | 30.22 ± 1.23 | >100 | 10.65 ± 0.97 |
| 20c | -OCH3 | -CH3 | 26.83 ± 1.22 | 23.35 ± 1.12 | >100 | 4.94 ± 0.64 |
| 20d | -OCH3 | Tert-Butyl | 33.24 ± 1.26 | 32.12 ± 1.18 | >100 | 10.46 ± 0.98 |
| 21a | -Br | -F | 45.68 ± 1.66 | 58.98 ± 1.76 | >100 | 37.59 ± 1.57 |
| 21b | -Br | -Cl | 11.13 ± 1.04 | 71.11 ± 1.85 | >100 | 13.78 ± 1.14 |
| 21c | -Br | -CH3 | 31.05 ± 1.49 | 22.74 ± 1.35 | >100 | 16.63 ± 1.22 |
| 21d | -Br | Tert-Butyl | 16.97 ± 1.22 | 16.16 ± 1.20 | >100 | 52.67 ± 1.72 |
| 22a | -3-Cl-4-F | -F | 19.63 ± 0.18 | 12.02 ± 0.91 | 52.52 ± 1.03 | 22.83 ± 1.35 |
| 22b | -3-Cl-4-F | -Cl | 6.443 ± 0.89 | >100 | 17.72 ± 0.95 | 6.84 ± 0.83 |
| 22c | -3-Cl-4-F | -CH3 | 11.274 ± 0.84 | >100 | 16.74 ± 0.88 | 16.81 ± 1.22 |
| 22d | -3-Cl-4-F | Tert-Butyl | 28.43 ± 0.89 | 22.40 ± 1.21 | 29.27 ± 2.23 | 46.30 ± 1.67 |
| 23a | -OCH3 | -Cl | 15.98 ± 1.10 | 39.76 ± 1.59 | >100 | 6.30 ± 0.79 |
| 23b | -OCH3 | Tert-Butyl | 2.71 ± 0.43 | 7.47 ± 0.87 | >100 | 6.55 ± 0.81 |
| 23c | -Br | -F | 32.12 ± 1.12 | 12.96 ± 0.84 | 39.76 ± 1.25 | 25.44 ± 1.43 |
| 23d | -3-Cl-4-F | -F | 6.08 ± 1.04 | 38.14 ± 0.96 | 57.85 ± 0.99 | 57.03 ± 1.75 |
| NVP-BEZ235 b | - | - | 0.54 ± 0.83 | 0.36 ± 0.86 | 0.20 ± 0.11 | 0.14 ± 0.10 |
| Sorafenib b | - | - | 3.97 ± 0.13 | 6.53 ± 0.23 | 3.03 ± 0.11 | 4.21 ± 0.15 |
| Compound | IC50(µM) | |
|---|---|---|
| PI3Kα | mTOR | |
| 19a | 0.72 | 2.62 |
| 23b | >10 | >10 |
| NVP-BEZ235 | 0.004 ± 0.002 | 0.006 ± 0.003 |
| PI101 | 0.011 ± 0.002 | 0.019 ± 0.002 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Lei, F.; Wang, C.; Zhang, B.; Yang, Z.; Li, W.; Zhu, W.; Xu, S. Design, Synthesis and Biological Evaluation of Novel Phenylsulfonylurea Derivatives as PI3K/mTOR Dual Inhibitors. Molecules 2018, 23, 1553. https://doi.org/10.3390/molecules23071553
Zhao B, Lei F, Wang C, Zhang B, Yang Z, Li W, Zhu W, Xu S. Design, Synthesis and Biological Evaluation of Novel Phenylsulfonylurea Derivatives as PI3K/mTOR Dual Inhibitors. Molecules. 2018; 23(7):1553. https://doi.org/10.3390/molecules23071553
Chicago/Turabian StyleZhao, Bingbing, Fei Lei, Caolin Wang, Binliang Zhang, Zunhua Yang, Wei Li, Wufu Zhu, and Shan Xu. 2018. "Design, Synthesis and Biological Evaluation of Novel Phenylsulfonylurea Derivatives as PI3K/mTOR Dual Inhibitors" Molecules 23, no. 7: 1553. https://doi.org/10.3390/molecules23071553







