Influence of Glutaraldehyde Cross-Linking Modes on the Recyclability of Immobilized Lipase B from Candida antarctica for Transesterification of Soy Bean Oil
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Magnetic Nanoparticles
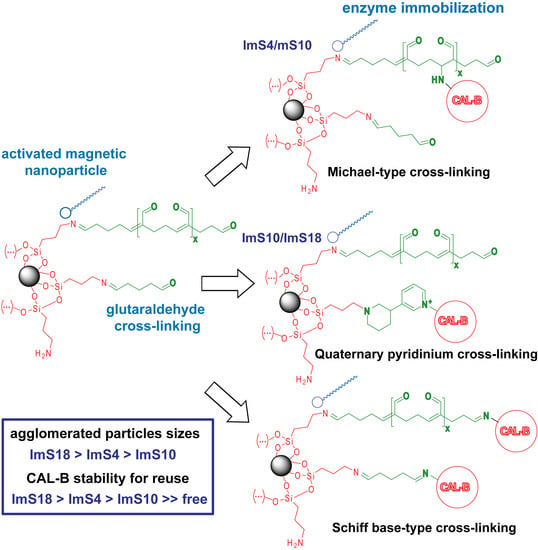
2.2. Improvement of CAL-B Immobilization Protocol
2.3. Immobilized CAL-B Characterization
2.4. Activity and Reuse Stability of Immobilized CAL-B for Biodiesel Synthesis
3. Materials and Methods
3.1. Materials
3.2. Characterization Techniques
3.3. Synthesis of Alkylimidazoles
3.4. Synthesis of ImS-n Sufactants
3.5. Magnetic Nanoparticles Synthesis
3.6. Nanoparticles Functionalization with Silanol Groups
3.7. Immobilization of Lipase B from Candida antarctica on Fe3O4@APTES
3.8. Lipase Catalytic Activity Optimization
3.9. Microbial Lipid Extraction Process
3.10. Biodiesel Synthesis Catalyzed by Lipase B from Candida antarctica Immobilized on Magnetic Nanoparticles
3.11. Determination of the Transesterification Reaction Activation Energy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Contesini, F.J.; Figueira, J.D.A.; Kawaguti, H.Y.; Fernandes, P.C.D.B.; Carvalho, P.D.O.; Nascimento, M.D.G.; Sato, H.H. Potential applications of carbohydrases immobilization in the food industry. Int. J. Mol. Sci. 2013, 14, 1335–1369. [Google Scholar] [CrossRef] [PubMed]
- Sirisha, V.L.; Jain, A.; Jain, A. Enzyme immobilization: An overview on methods, support material, and applications of immobilized enzymes. Adv. Food Nutr. Res. 2016, 79, 179–211. [Google Scholar] [PubMed]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netto, C.G.C.M.; Toma, H.E.; Andrade, L.H. Superparamagnetic nanoparticles as versatile carriers and supporting materials for enzymes. J. Mol. Catal. B-Enzym. 2013, 85–86, 71–92. [Google Scholar] [CrossRef]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. A general overview of support materials for enzyme immobilization: Characteristics, properties, practical utility. Catalysts 2018, 8, 92. [Google Scholar] [CrossRef]
- Vaghari, H.; Jafarizadeh-Malmiri, H.; Mohammadlou, M.; Berenjian, A.; Anarjan, N.; Jafari, N.; Nasiri, S. Application of magnetic nanoparticles in smart enzyme immobilization. Biotechnol. Lett. 2016, 38, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.A.; Park, H.J.; Driscoll, A.J. Enzyme nanoparticle fabrication: Magnetic nanoparticle synthesis and enzyme immobilization. Methods Mol. Biol. 2011, 679, 183–191. [Google Scholar] [PubMed]
- Mikhaylova, M.; Kim, D.K.; Bobrysheva, N.; Osmolowsky, M.; Semenov, V.; Tsakalakos, T.; Muhammed, M. Superparamagnetism of magnetite nanoparticles: Dependence on surface modification. Langmuir 2004, 20, 2472–2477. [Google Scholar] [CrossRef] [PubMed]
- Al-Qodah, Z.; Al-Shannag, M.; Al-Busoul, M.; Penchev, I.; Orfali, W. Immobilized enzymes bioreactors utilizing a magnetic field: A review. Biochem. Eng. J. 2017, 121, 94–106. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Lotti, M.; Pleiss, J.; Valero, F.; Ferrer, P. Enzymatic production of biodiesel: Strategies to overcome methanol inactivation. Biotechnol. J. 2018, 13, 1700155. [Google Scholar] [CrossRef] [PubMed]
- Mehrasbi, M.R.; Mohammadi, J.; Peyda, M.; Mohammadi, M. Covalent immobilization of candida antarctica lipase on core-shell magnetic nanoparticles for production of biodiesel from waste cooking oil. Renew. Energ. 2017, 101, 593–602. [Google Scholar] [CrossRef]
- Ferrario, V.; Veny, H.; Angelis, E.D.; Navarini, L.; Ebert, C.; Gardossi, L. Lipases immobilization for effective synthesis of biodiesel starting from coffee waste oils. Biomolecules 2013, 3, 514–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Li, X.; He, T. Kinetics of (3-aminopropyl)triethoxylsilane (aptes) silanization of superparamagnetic iron oxide nanoparticles. Langmuir 2013, 29, 15275–15282. [Google Scholar] [CrossRef] [PubMed]
- Farrel, D.; Majetich, S.; Wilcoxon, J. Preparation and characterization of monodisperse Fe nanoparticles. J. Phys. Chem. B 2003, 107, 11022–11030. [Google Scholar] [CrossRef]
- Okuda, K.; Urabe, I.; Yamada, Y.; Okada, H. Reaction of glutaraldehyde with amino and thiol compounds. J. Ferment. Bioeng. 1991, 71, 100–105. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques 2004, 37, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Weetall, H.H. Immobilized enzymes: Analytical applications. Anal. Chem. 1974, 46, 602A–615A. [Google Scholar] [PubMed]
- Richards, F.M.; Knowles, J.R. Glutaraldehyde as a protein cross-linkage reagent. J. Mol. Biol. 1968, 37, 231–233. [Google Scholar] [CrossRef]
- Hardy, P.M.; Hughes, G.J.; Rydon, H.N. The nature of the cross-linking of proteins by glutaraldehyde. Part 2.L the formation of quaternary pyridinium compounds by the action of glutaraldehyde on proteins and the identification of a 3-(2-piperidyl)-pyridinium derivative, anabilysine, as a cross-linking entity. J. Chem. Soc. Perkin Trans. 1 1979, 0, 2282–2288. [Google Scholar]
- Yi, S.; Dai, F.; Zhao, C.; Si, Y. A reverse micelle strategy for fabricating magnetic lipase immobilized nanoparticles with robust enzymatic activity. Sci. Rep. 2017, 7, 9806. [Google Scholar] [CrossRef] [PubMed]
- Souza, F.D.S.; B, S.; Tondo, D.W.; Leopoldino, E.C.; Fiedler, H.D.; Nome, F. Imidazolium-based zwitterionic surfactants: Characterization of normal and reverse micelles and stabilization of nanoparticles. Langmuir 2015, 31, 3587–3595. [Google Scholar] [CrossRef] [PubMed]
- Salviano, L.B.; Cardoso, T.M.D.S.; Silva, G.C.; Dantas, M.S.S.; Ferreira, A.D.M. Microstructural assessment of magnetite nanoparticles (Fe3O4) obtained by chemical precipitation under different synthesis conditions. Mater. Res. 2018, 21, e20170764. [Google Scholar] [CrossRef]
- Lim, J.; Yeap, S.P.; Che, H.X.; Low, S.C. Characterization of magnetic nanoparticle by dynamic light scattering. Nanoscale Res. Lett. 2013, 8, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lorente, G.; Palomo, J.M.; Mateo, C.; Munilla, R.; Ortiz, C.; Cabrera, Z.; Guisan, J.M.; Fernandez-Lafuente, R. Glutaraldehyde cross-linking of lipases adsorbed on aminated supports in the presence of detergents leads to improved performance. Biomacromolecules 2006, 7, 2610–2615. [Google Scholar] [CrossRef] [PubMed]
- Nicolás, P.; Lassalle, V.L.; Ferreira, M.L. Quantification of immobilized candida antarctica lipase b (calb)using icp-aes combined with bradford method. Enzyme Microb. Technol. 2017, 97, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, H.; Liu, W.; Wang, N.; Yu, X. Improved performance of magnetic cross-linked lipase aggregates by interfacial activation: A robust and magnetically recyclable biocatalyst for transesterification of jatropha oil. Molecules 2017, 22, E2157. [Google Scholar] [CrossRef] [PubMed]
- Vršanská, M.; Voberková, S.; Jiménez, A.M.J.; Strmiska, V.; Adam, V. Preparation and optimisation of cross-linked enzyme aggregates using native isolate white rot fungi trametes versicolor and fomes fomentarius for the decolourisation of synthetic dyes. Int. J. Environ. Res. Public Health 2018, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.W.; Shaw, J.F.; Yang, K.H.; Chang, S.F.; Shieh, C.J. Studies of optimum conditions for covalent immobilization of candida rugosa lipase on poly(γ-glutamic acid) by rsm. Biores. Technol. 2008, 99, 2800–2805. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.S.; Long, S.Y.; Huang, J.; Xiao, H.Y.; Zhou, J.Y. Immobilization of pycnoporus sanguineus laccase on magnetic chitosan microspheres. Biochem. Eng. J. 2005, 25, 15–23. [Google Scholar] [CrossRef]
- Imai, K.; Shiomi, T.; Uchida, K.; Miya, M. Immobilization of enzyme onto poly(ethy1ene-vinyl alcohol) membrane. Biotechnol. Bioeng. 1986, XXVIII, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Grazu, V.; Abian, O.; Mateo, C.; Batista-Viera, F.; Fernandez-Lafuente, R.; Guisa´n, J.M. Stabilization of enzymes by multipoint immobilization of thiolated proteins on new epoxy-thiol supports. Biotechnol. Bioeng. 2005, 90, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.C.; Lia, Y.K. Application of bacterial cellulose pellets in enzyme immobilization. J. Mol. Catal. B-Enzym. 2008, 54, 103–108. [Google Scholar] [CrossRef]
- Netto, C.G.C.M.; Nakamura, M.; Andrade, L.H.; Toma, H.E. Improving the catalytic activity of formate dehydrogenase from candida boidinii by using magnetic nanoparticles. J. Mol. Catal. B-Enzym. 2012, 84, 136–143. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R.; Rodrıguez, V.; Mateo, C.; Penzol, G.; Hernandez-Justiz, O.; Irazoqui, G.; Villarino, A.; Ovsejevi, K.; Batista, F.; Guisan, J.M. Stabilization of multimeric enzymes via immobilization and post-immobilization techniques. J. Mol. Catal. B-Enzym. 1999, 7, 181–189. [Google Scholar] [CrossRef]
- Pedroche, J.; Yust, M.d.M.; Mateo, C.; Fernandez-Lafuente, R.; Giron-Calle, J.; Alaiz, M.; Vioque, J.; Guisan, J.M.; Millan, F. Effect of the support and experimental conditions in the intensity of the multipoint covalent attachment of proteins on glyoxyl-agarose supports: Correlation between enzyme–support linkages and thermal stability. Enzyme Microb. Technol. 2007, 40, 1160–1166. [Google Scholar] [CrossRef]
- Song, J.E.; Song, W.S.; Yeo, S.Y.; Kim, H.R.; Lee, S.H. Covalent immobilization of enzyme on aminated woven poly (lactic acid) via ammonia plasma: Evaluation of the optimum immobilization conditions. Text. Res. J. 2016, 87, 1177–1191. [Google Scholar] [CrossRef]
- Feng, J.; Yu, S.; Li, J.; Mo, T.; Li, P. Enhancement of the catalytic activity and stability of immobilized aminoacylase using modified magnetic Fe3O4 nanoparticles. Chem. Eng. J. 2016, 286, 216–222. [Google Scholar] [CrossRef]
- Zhu, J.; Sun, G. Lipase immobilization on glutaraldehyde-activated nanofibrous membranes for improved enzyme stabilities and activities. React. Funct. Polym. 2012, 72, 839–845. [Google Scholar] [CrossRef]
- Dunwell, M.; Yan, Y.; Xu, B. In situ infrared spectroscopic investigations of pyridine-mediated CO2 reduction on pt electrocatalysts. ACS Catal. 2017, 7, 5410–5419. [Google Scholar] [CrossRef]
- Monsan, P.; Puzo, G.; Mazarguil, H. Étude du mécanisme d’établissement des liaisons glutaraldehyde protéines. Biochime 1975, 57, 1281–1292. [Google Scholar] [CrossRef]
- Betancor, L.; Lopez-Gallego, F.; Hidalgo, A.; Alonso-Morales, N.; Dellamora-Ortiz, G.; Mateo, C.; Fernandez-Lafuente, R.; Guisan, J.M. Different mechanisms of protein immobilization on glutaraldehyde activated supports: Effect of support activation and immobilization conditions. Enzyme Microb. Technol. 2006, 39, 877–882. [Google Scholar] [CrossRef]
- Priebe, J.P.; Souza, F.D.; Silva, M.; Tondo, D.W.; Priebe, J.M.; Micke, G.A.; Costa, A.C.O.; Bunton, C.A.; Quina, F.H.; Fiedler, H.D.; et al. The chameleon-like nature of zwitterionic micelles: Effect of cation binding. Langmuir 2012, 28, 1758–1764. [Google Scholar] [CrossRef] [PubMed]
- Drinkel, E.D.; Souza, F.D.; Fiedler, H.; Nome, F. The chameleon effect in zwitterionic micelles: Binding of anions and cations and use as nanoparticle stabilizing agents. Curr. Opin. Colloid Interface Sci. 2013, 18, 26–34. [Google Scholar] [CrossRef]
- Peters, K.; Richards, F.M. Chemical cross-linking: Reagents and problems in studies of membrane structure. Ann. Rev. Biochem. 1977, 46, 523–551. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Mauguen, Y.; Prangé, T. Revisiting glutaraldehyde cross-linking: The case of the arg–lys intermolecular doublet. Acta. Cryst. 2010, F66, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Habeeb, A.F.S.A.; Hiramoto, R. Reaction of proteins with glutaraldehyde. Arch. Biochemi. Biophys. 1968, 126, 16–26. [Google Scholar] [CrossRef]
- Kuznetsova, N.P.; Mishaeva, R.N.; Gudkin, L.R.; Panarin, E.F. Reactions of glutaraldehyde with dipolar ions of amino acids and proteins. Russ. Chem. Bll. 2013, 62, 918–927. [Google Scholar] [CrossRef]
- Costa, V.M.; Souza, M.C.M.D.; Fechine, P.B.A.; Macedo, A.C.; Gonçalves, L.R.B. Nanobiocatalytic systems based on lipase-fe3o4 and conventional systems for isoniad synthesis: A comparative study. Braz. J. Chem. Eng. 2016, 33, 661–673. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. The slow-down of the calb immobilization rate permits to control the inter and intra molecular modification produced by glutaraldehyde. Proc. Biochem. 2012, 47, 766–774. [Google Scholar] [CrossRef]
- Katcka, M.; Urbanski, T. Infrared absorption spectra of quaternary salts of pyridine. Bull. Acad. Pol. Sci. 1964, XII, 615–621. [Google Scholar]
- Bowes, J.H.; Cater, C.W. The reaction of glutaraldehyde with proteins and other biological materials. J. R. Microsc. Soc. 1966, 85, 193–200. [Google Scholar] [CrossRef]
- Roduner, E. Understanding catalysis. Chem. Soc. Rev. 2014, 43, 8226–8239. [Google Scholar] [CrossRef] [PubMed]
- Busto, M.D.; Apenten, R.K.O.; Robinson, D.S.; Wu, Z.; Casey, R.; Hughes, R.K. Kinetics of thermal inactivation of pea seed lipoxygenases and the effect of additives on their thermostability. Food Chem. 1999, 65, 323–329. [Google Scholar] [CrossRef]
- Kazemi, M.; Himo, F.; Åqvist, J. Enzyme catalysis by entropy without circe effect. Proc. Natl. Acad. Sci. USA 2016, 113, 2406–2411. [Google Scholar] [CrossRef] [PubMed]
- Åqvist, J.; Kazemi, M.; Isaksen, G.V.; Brandsdal, B.O. Entropy and enzyme catalysis. Acc. Chem. Res. 2017, 50, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Brask, J.; Mohammadi, M. Preparation of highly reusable biocatalysts by immobilization of lipases on epoxy-functionalized silica for production of biodiesel from canola oil. Biochem. Eng. J. 2015, 101, 23–31. [Google Scholar] [CrossRef]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Chen, B.; Miller, E.M.; Miller, L.; Maikner, J.J.; Gross, R.A. Effects of macroporous resin size on candida antarctica lipase b adsorption, fraction of active molecules, and catalytic activity for polyester synthesis. Langmuir 2007, 23, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Alagöz, D.; Tükel, S.S.; Yildirim, D. Immobilization of pectinase on silica-based supports: Impacts ofparticle size and spacer arm on the activity. Int. J. Biol. Macromol. 2016, 87, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, G.; Tao, K.; Feng, M.; Zhao, X.; Li, Z.; Xu, H.; Xia, D.; Lu, J.R. Immobilization of lipases on alkyl silane modified magnetic nanoparticles: Effect of alkyl chain length on enzyme activity. PLoS ONE 2012, 7, e43478. [Google Scholar] [CrossRef] [PubMed]
- Venuti, E.; Shishmarev, D.; Kuchel, P.W.; Dutt, S.; Blumenthal, C.S.; Gaskin, K.J. Bile salt stimulated lipase: Inhibition by phospholipids and relief by phospholipase A2. J. Cyst. Fibros. 2017, 16, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Yamaura, M.; Camilo, R. L.; Sampaio, L.C.; Macedo, M.A.; Nakamura, M.; Toma, H.E. Preparation and characterization of (3-aminopropyl) triethoxysilane coated magnetite nanoparticles. J. Magn. Mater. 2004, 279, 210–217. [Google Scholar] [CrossRef]
- Thangaraj, B.; Zhaohua, J.; Lingmei, D.; Dehua, L.; Wi, D. Lipase NS81006 immobilized on Fe3O4 magnetic nanoparticles for biodiesel production. Ovid. Univ. Press 2016, 27, 13–21. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.J.; Petricevic, S.F.; Coddington, J.M.; Stanley, R.A. AN NMR assay for quantitating lipase activity in biphasic macroemulsions. J. Am. Oil Chem. Soc. 1992, 69, 295–300. [Google Scholar]
- Eyring, H. The activated complex in chemical reactions. J. Chem. Phys. 1935, 3, 107–115. [Google Scholar] [CrossRef]
Sample Availability: Not available. |







| Biocatalyst | Ea (kJ mol−1) | ΔS ≠ (J mol−1) | k (μMs−1) a | Yield (%) b |
|---|---|---|---|---|
| Free CAL-B | 29.4 | −73 | 0.048 | 2/1.5 c |
| NP-ImS4/CAL-B | 25.6 | −138 | 33.2 | 79.7 |
| NP-ImS10/CAL-B | 18.2 | −142 | 21.7 | 90 |
| NP-ImS18/CAL-B | 27.8 | −145 | 14.0 | 83 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
A. Modenez, I.; Sastre, D.E.; C. Moraes, F.; Marques Netto, C.G.C. Influence of Glutaraldehyde Cross-Linking Modes on the Recyclability of Immobilized Lipase B from Candida antarctica for Transesterification of Soy Bean Oil. Molecules 2018, 23, 2230. https://doi.org/10.3390/molecules23092230
A. Modenez I, Sastre DE, C. Moraes F, Marques Netto CGC. Influence of Glutaraldehyde Cross-Linking Modes on the Recyclability of Immobilized Lipase B from Candida antarctica for Transesterification of Soy Bean Oil. Molecules. 2018; 23(9):2230. https://doi.org/10.3390/molecules23092230
Chicago/Turabian StyleA. Modenez, Iago, Diego E. Sastre, Fernando C. Moraes, and Caterina G. C. Marques Netto. 2018. "Influence of Glutaraldehyde Cross-Linking Modes on the Recyclability of Immobilized Lipase B from Candida antarctica for Transesterification of Soy Bean Oil" Molecules 23, no. 9: 2230. https://doi.org/10.3390/molecules23092230
APA StyleA. Modenez, I., Sastre, D. E., C. Moraes, F., & Marques Netto, C. G. C. (2018). Influence of Glutaraldehyde Cross-Linking Modes on the Recyclability of Immobilized Lipase B from Candida antarctica for Transesterification of Soy Bean Oil. Molecules, 23(9), 2230. https://doi.org/10.3390/molecules23092230