Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O?-Acac)(?-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Vitro cytotoxicity of Ptac2S Complex in SKOV-3 Cells
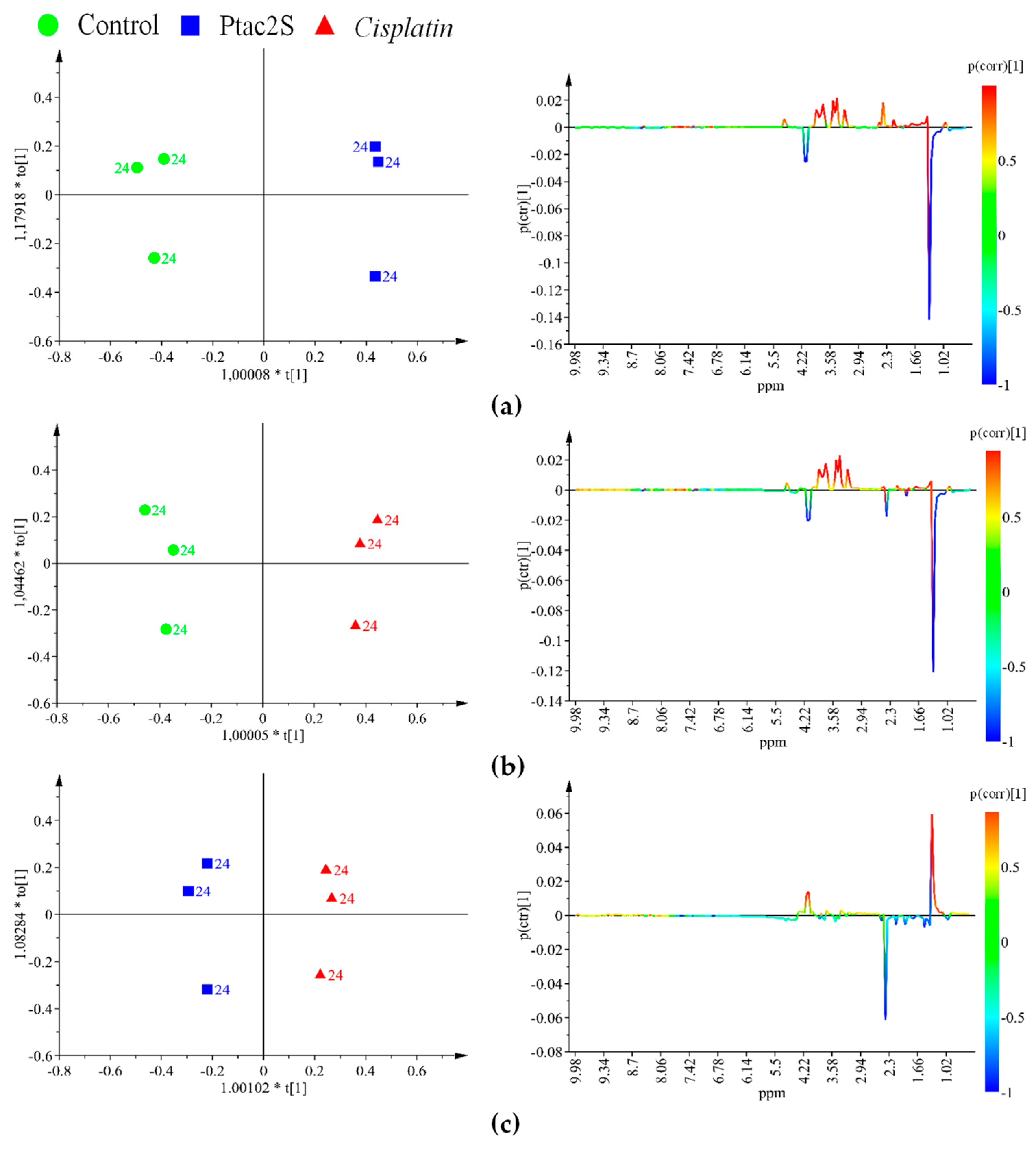
2.2. Metabolic Alterations in SKOV-3 Cells Induced by Ptac2S Treatment
2.2.1. Metabolomic Studies of the Aqueous Skov-3 Cells Extracts.
2.2.2. Metabolomic Studies of Lipidic Skov-3 Cells Extracts.
2.3. Metabolic Changes of Skov-3 Cells Culture Media Due to Treatment with Ptac2s
3. Materials and Methods
3.1. Synthesis of Complexes
3.2. SKOV-3 Cell Cultures and Drugs Administration
3.2.1. Cytotoxicity Assay
3.2.2. Metabolomic Assay.
3.3. NMR Measurements
3.4. Spectral Processing and Multivariate Data Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beger, R.D. A Review of Applications of Metabolomics in Cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Rubayet Hasan, M. Cancer Metabolism and Drug Resistance. Metabolites 2015, 5, 571–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, Y.L.; Griffiths, J.R. Using metabolomics to monitor anticancer drugs. Ernst Schering Found. Symp. Proc. 2007, 4, 55–78. [Google Scholar]
- Vander Heiden, M.G. Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 2011, 10, 671–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcìa-Alvarez, I.; Fernandez-Mayoralas, A.; Garrido, L. Effect of drugs in cells and tissue by NMR spectroscopy. Curr. Top. Med. Chem. 2011, 11, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.L.; Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer 2004, 4, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Maruvada, P.; Milner, J.A. Metabolomics in biomarker discovery: Future uses for cancer prevention. Future Oncol. 2008, 4, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rainaldi, G.; Romano, R.; Indovina, P.; Ferrante, A.; Motta, A.; Indovina, P.L.; Santini, M.T. Metabolomics Using 1H-NMR of Apoptosis and Necrosis in HL60 Leukemia Cells: Differences between the Two Types of Cell Death and Independence from the Stimulus of Apoptosis Used. Radiat. Res. 2008, 169, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Triba, M.N.; Starzec, A.; Bouchemal, N.; Guenin, E.; Perret, G.Y.; Moyec, L.L. Metabolomic profiling with NMR discriminates between biphosphonate and doxorubicin effects on B16 melanoma cells. NMR Biomed. 2010, 23, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Bayet-Robert, M.; Morvan, D.; Chollet, P.; Barthomeuf, C. Pharmacometabolomics of Docetaxel-treated Human MCF-7 breast cancer cells provides evidence of varying cellular responses at high and low doses. Breast Cancer Res. Treat. 2010, 120, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Bayet-Robert, M.; Loiseau, D.; Rio, P.; Demidem, A.; Barthomeuf, C.; Stepien, G.; Morvan, D. Quantitative two-dimensional HRMAS 1H-NMR spectroscopy-based metabolite profiling of human cancer cell lines and response to chemotherapy. Magn. Reson. Med. 2010, 63, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Klawitter, J.; Anderson, N.; Klawitter, J.; Christians, U.; Leibfritz, D.; Eckhardt, S.G.; Serkova, N.J. Time-Dependent Effects of Imatinib in Human Leukaemia Cells: A Kinetic NMR-Profiling Study. Br. J. Cancer. 2009, 100, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Lodi, A.; Ronen, S.M. Magnetic resonance spectroscopy detectable metabolomic fingerprint of response to antineoplastic treatment. PLoS ONE 2011, 6, e26155. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.F.; Ladeirinha, A.F.; Lamego, I.; Gil, A.M.; Carvalho, L.; Carreira, I.M.; Melo, J.B. Potential Markers of Cisplatin Treatment Response Unveiled by NMR Metabolomics of Human Lung Cells. Mol. Pharm. 2013, 10, 4242–4251. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tong, Y.; Wang, J.; Huang, Y. NMR Studies of the relationship between the changes of membrane lipids and the cisplatin-resistance of A549/DDP Cells. Cancer Cell Int. 2003. [Google Scholar] [CrossRef]
- Duarte, I.F.; Lamego, I.; Marques, J.; Marques, M.P.M.; Blaise, B.J.; Gil, A.M. Nuclear magnetic resonance (NMR) study of the effect of cisplatin on the metabolic profile of MG-63 osteosarcoma cells. J. Proteome Res. 2010, 9, 5877–5886. [Google Scholar] [CrossRef] [PubMed]
- Mirbahai, L.; Wilson, M.; Shaw, C.S.; McConville, C.; Malcomson, R.D.G.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. 1H Magnetic resonance spectroscopy metabolites as biomarkers for cell cycle arrest and cell death in rat glioma cells. Int. J. Biochem. Cell Biol. 2011, 43, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Mirbahai, L.; Wilson, M.; Shaw, C.S.; McConville, C.; Malcomson, R.D.G.; Kauppinen, R.A.; Peet, A.C. Lipid biomarkers of glioma cell growth arrest and cell death detected by 1H Magic Angle Spinning MRS. NMR Biomed. 2012, 25, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wilson, M.; Mirbahai, L.; McConville, C.; Arvanitis, T.N.; Griffin, J.L.; Kauppinen, R.A.; Peet, A.C. In vitro metabonomic study detects increases in UDP-GlcNAc and UDP-GalNAc, as early markers of cisplatin treatment response in brain tumor cells. J. Proteome Res. 2011, 10, 3493–3500. [Google Scholar] [CrossRef] [PubMed]
- De Pascali, S.A.; Papadia, P.; Capoccia, S.; Marchio, L.; Lanfranchi, M.; Ciccarese, A.; Fanizzi, F.P. Hard/soft selectivity in ligand substitution reactions of beta-diketonate platinum (II) complexes. Dalton Trans. 2009, 37, 7786–7795. [Google Scholar] [CrossRef] [PubMed]
- Muscella, A.; Calabriso, N.; De Pascali, S.A.; Urso, L.; Ciccarese, A.; Fanizzi, F.P.; Migoni, D.; Marsigliante, S. New platinum (II) complexes containing both an O,O′-chelated acetylacetonate ligand and a sulfur ligand in the platinum coordination sphere induce apoptosis in HeLa cervical carcinoma cells. Biochem. Pharmacol. 2007, 74, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Muscella, A.; Calabriso, N.; De Pascali, S.A.; Urso, L.; Ciccarese, A.; Fanizzi, F.P.; Migoni, D.; Marsigliante, S. [Pt(O,O′-acac)(γ-acac)(DMS)], a new Pt compound exerting fast cytotoxicity in MCF-7 breast cancer cells via the mitochondrial apoptotic pathway. Br. J. Pharmacol. 2008, 153, 34–39. [Google Scholar] [CrossRef] [PubMed]
- De Pascali, S.A.; Papadia, P.; Ciccarese, A.; Pacifico, C.; Fanizzi, F.P. First examples of β-diketonate platinum II complexes with sulfoxide ligands. Eur. J. Inorg. Chem. 2005, 5, 788–796. [Google Scholar] [CrossRef]
- De Pascali, S.A.; Lugoli, F.; De Donno, A.; Fanizzi, F.P. Mutagenic tests confirm that new acetylacetonate Pt(II) complexes induce apoptosis in cancer cells interacting with nongenomic biological targets. Metal-Based Drugs 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Marzo, T.; De Pascali, S.A.; Gabbiani, C.; Fanizzi, F.P.; Messori, L.; Pratesi, A. ESI-MS studies of the reactions of novel platinum(II) complexes containing O,O′-chelated acetylacetonate and sulfur ligands with selected model proteins. Biometals 2017, 30, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Mezencev, R. Interactions of cisplatin with non-DNA targets and their influence on anticancer activity and drug toxicity: The complex world of the platinum complex. Curr. Cancer Drug Targets 2015, 14, 794–816. [Google Scholar] [CrossRef]
- Florea, A.M.; Büsselberg, D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed]
- Arnesano, F.; Banci, L.; Bertini, I.; Felli, I.C.; Losacco, M.; Natile, G. Probing the interaction of cisplatin with the human copper chaperone Atox1 by solution and in-cell NMR spectroscopy. J. Am. Chem. Soc. 2011, 133, 18361–18369. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Benedetti, M.; Natile, G.; Marzilli, L.G. Basic coordination chemistry relevant to DNA adducts formed by the cisplatin anticancer drug. NMR studies on compounds with sterically crowded chiral ligands. Inorg. Chem. 2010, 49, 5573–5583. [Google Scholar] [CrossRef] [PubMed]
- Saad, J.S.; Benedetti, M.; Natile, G.; Marzilli, L.G. NMR Studies of Models Having the Pt(d(GpG)) 17-Membered Macrocyclic Ring Formed in DNA by Platinum Anticancer Drugs: Pt Complexes with Bulky Chiral Diamine Ligands. Inorg. Chem. 2011, 50, 4559–4571. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Saad, J.S.; Marzilli, L.G.; Natile, G. Chiral Discrimination in the formation reaction and at equilibrium for N,N,N’,N’-tetramethyl-1,2-diaminocyclohexane-PtG2 Complexes. Dalton Trans. 2003, 5, 872–879. [Google Scholar] [CrossRef]
- Benedetti, M.; Tamasi, G.; Cini, R.; Natile, G. X-ray Structure and Circular Dichroism of Pure Rotamers of Bis[guanosine-5′-monophosphate(-1)](N,N,N’,N’-tetramethylcyclohexyl-1,2-diamine) platinum(ii) Complexes That Have R,R and S,S Configurations at the Asymmetric Diamine. Chem. Eur. J. 2003, 9, 6122–6132. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Marzilli, L.G.; Natile, G. Rotamer Stability in cis-[Pt(diA)G2] Complexes (diA = Diamine Derivative and G = Guanine Derivative) Mediated by Carrier-Ligand Amine Stereochemistry as Revealed by Circular Dichroism Spectroscopy. Chem. Eur. J. 2005, 11, 5302–5310. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Tamasi, G.; Cini, R.; Marzilli, L.G. The First Pure HT Rotamer of a Complex with a cis-[Metal(nucleotide)2] Unit: A cis-[Pt(amine)2(nucleotide)2] HT Rotamer with Unique Molecular Structural Features. Chem. Eur. J. 2007, 13, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Ducani, C.; Migoni, D.; Antonucci, D.; Vecchio, V.M.; Ciccarese, A.; Romano, A.; Verri, T.; Ciccarella, G.; Fanizzi, F.P. Experimental Evidence That a DNA Polymerase Can Incorporate N7-Platinated Guanines To Give Platinated DNA. Angew. Chem. Int. Ed. 2008, 47, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Romano, A.; De Castro, F.; Girelli, C.R.; Antonucci, D.; Migoni, D.; Verri, T.; Fanizzi, F.P. N7-platinated ribonucleotides are not incorporated by RNA polymerases. New perspectives for a rational design of platinum antitumor drugs. J. Inorg. Biochem. 2016, 163, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Carrisi, C.; Antonucci, D.; Lunetti, P.; Migoni, D.; Girelli, C.R.; Dolce, V.; Fanizzi, F.P.; Benedetti, M.; Capobianco, L. Transport of platinum bonded nucleotides into proteoliposomes, mediated by Drosophila melanogaster thiamine pyrophosphate carrier protein (DmTpc1). J. Inorg. Biochem. 2014, 130, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Lunetti, P.; Romano, A.; Carrisi, C.; Antonucci, D.; Verri, T.; De Benedetto, G.E.; Dolce, V.; Fanizzi, F.P.; Benedetti, M.; Capobianco, L. Platinated Nucleotides are Substrates for the Human Mitochondrial Deoxynucleotide Carrier (DNC) and DNA Polymerase γ: Relevance for the Development of New Platinum-Based Drugs. ChemistrySelect 2016, 1, 4633–4637. [Google Scholar] [CrossRef]
- Fan, T.W.; Lane, A.N.; Higashi, R.M. The promise of metabolomics in cancer molecular therapeutics. Curr. Opin. Mol. Ther. 2004, 6, 584–592. [Google Scholar] [PubMed]
- Muscella, A.; Vetrugno, C.; Fanizzi, F.P.; Manca, C.; De Pascali, S.A.; Marsigliante, S. A new platinum (II) compound anticancer drug candidate with selective cytotoxicity for breast cancer cells. Cell Death Dis. 2013, 4, e796. [Google Scholar] [CrossRef] [PubMed]
- Muscella, A.; Vetrugno, C.; Cossa, L.G.; Antonaci, G.; De Nuccio, F.; De Pascali, S.A.; Fanizzi, F.P.; Marsigliante, S. In Vitro and In Vivo Antitumor Activity of [Pt(O,O′-acac)(γ-acac)(DMS)] in Malignant Pleural Mesothelioma. PLoS ONE 2016, 11, e0165154. [Google Scholar] [CrossRef] [PubMed]
- Vetrugno, C.; Muscella, A.; Fanizzi, F.P.; Cossa, L.G.; Migoni, D.; De Pascali, S.A.; Marsigliante, S. Different apoptotic effects of [Pt(O,O′-acac)(γ-acac)(DMS)] and cisplatin on normal and cancerous human epithelial breast cells in primary culture. Br. J. Pharmacol. 2014, 171, 5139–5153. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.K.; Bundy, B.; Wenzel, L.; Huang, H.Q.; Baergen, R.; Lele, S.; Copeland, L.J.; Walker, J.L.; Burger, R.A. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 2006, 5, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Alberts, D.S.; Green, S.; Hannigan, E.V.; O′Toole, R.; Stock-Novack, D.; Anderson, P.; Surwit, E.A.; Malvlya, V.K.; Nahhas, W.A.; Jolles, C.J. Improved therapeutic index of carboplatin plus cyclophosphamide versus cisplatin plus cyclophosphamide: Final report by the Southwest Oncology Group of a phase III randomized trial in stages III and IV ovarian cancer. J. Clin. Oncol. 1992, 10, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Ozols, R.F.; Bundy, B.N.; Greer, B.E.; Fowler, J.M.; Clarke-Pearson, D.; Burger, R.A.; Mannel, R.S.; De Geest, K.; Hartenbach, E.M.; Baergen, R. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: A Gynecologic Oncology Group study. J. Clin. Oncol. 2003, 21, 3194–3200. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Smigal, C.; Thun, M.J. Cancer statistics, 2006. CA Cancer J. Clin. 2006, 56, 106–130. [Google Scholar] [CrossRef] [PubMed]
- Ben Sellem, D.; Elbayed, K.; Neuville, A.; Moussallieh, F.-M.; Lang-Averous, G.; Piotto, M.; Bellocq, J.-P.; Namer, I.J. Metabolomic Characterization of Ovarian Epithelial Carcinomas by HRMAS-NMR Spectroscopy. J. Oncol. 2011, 2011, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Li, J.; Wu, H.; Chen, Z. Metabolic responses of HeLa cells to silica nanoparticles by NMR based metabolomic analyses. Metabolomics 2013, 9, 874–886. [Google Scholar] [CrossRef]
- Tiziani, S.; Lodi, A.; Khanim, F.L.; Viant, M.R. Christopher M. Bunce, Ulrich L. Günther. Metabolomic Profiling of Drug Responses in Acute Myeloid Leukaemia Cell Lines. PLoS ONE 2009, 4, e4251. [Google Scholar] [CrossRef]
- Morvan, D.; Demidem, A. Metabolomics by Proton Nuclear Magnetic Resonance Spectroscopy of the Response to Chloroethylnitrosourea Reveals Drug Efficacy and Tumor Adaptive Metabolic Pathways. Cancer Res. 2007, 67, 2150–2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmnas, M.S.A.; Vogel, H.J. The Future of NMR Metabolomics in Cancer Therapy: Towards Personalizing Treatment and Developing Targeted Drugs? Metabolites 2013, 3, 373–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbet, C.; Olivier, F. Cancer cell metabolism and mitochondria: Nutrient plasticity for TCA cycle fueling. Bioch. Bioph. Acta 2017, 1868, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R. Lactate dehydrogenase 5: An old friend and a new hope in the war on cancer. Cancer Lett. 2015, 358, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mingeot-Leclercq, M.P.; Brasseur, R.; Schanck, A. Molecular parameters involved in aminoglycoside nephrotoxicity. J. Toxicol. Environ Health 1995, 44, 263–300. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, F.G.; Storrs, R.W.; Naumovski, L.; Goralski, T.; Spielman, D. Detection of apoptotic cell death by proton nuclear magnetic resonance spectroscopy. Blood 1996, 87, 1951–1956. [Google Scholar] [PubMed]
- Mikhailenko, V.M.; Philchenkov, A.A.; Zavelevich, M.P. Analysis of 1H-NMR-detectable domains for assessment of apoptosis induced by inhibitors of DNA synthesis and replication. Cell Biol. Int. 2005, 29, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Podo, F. Tumour phospholipid metabolism. NMR Biomed. 1999, 12, 413–439. [Google Scholar] [CrossRef]
- Michel, V.; Ramsubir, Z.Y.S.; Bakovic, M. Choline transport for phospholipids synthesis. Exp. Biol. Med. 2006, 231, 490–504. [Google Scholar] [CrossRef]
- Anthony, M.L.; Zhao, M.; Brindle, K.M. Inhibition of phosphatidylcholine biosynthesis following induction of apoptosis in HL60 cells. J. Biol. Chem. 1999, 274, 19686–19692. [Google Scholar] [CrossRef] [PubMed]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.L.; Lehtimaki, K.K.; Valonen, P.K.; Grohn, O.H.J.; Kettunen, M.I.; Yla-Herttuala, S.; Pitkanen, A.; Nicholson, J.K.; Kauppinen, R.A. Assignment of 1H Nuclear Magnetic Resonance Visible Polyunsaturated Fatty Acids in BT4C Gliomas Undergoing Ganciclovir-Thymidine Kinase Gene Therapy-Induced Programmed Cell Death. Cancer Res. 2003, 63, 3195–3201. [Google Scholar] [PubMed]
- Delikatny, E.J.; Chawla, S.; Leung, D.-J.; Poptani, H. MR Visible Lipids and the Tumour Microenvironment. NMR Biomed. 2011, 24, 592–611. [Google Scholar] [PubMed]
- Zietkowski, D.; Payne, G.S.; Nagy, E.; Mobberley, M.A.; Ryder, T.A.; De Souza, N.M. Comparison of NMR Lipid Profiles in Mitotic Arrest and Apoptosis as Indicators of Paclitaxel Resistance in Cervical Cell Lines. Magn. Reson. Med. 2012, 68, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.-L.; Hu, X.; Tam, K.Y. Targeting Tumor Metabolism for Cancer Treatment: Is Pyruvate Dehydrogenase Kinases (PDKs) a Viable Anticancer Target? Int. J. Biol. Sci. 2015, 11, 1390–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhara, S.C. Rapid method for the synthesis of cis-[Pt(NH3)2Cl2]. Indian J. Chem. 1970, 8, 193–194. [Google Scholar]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.-M. Metabolite profiling by one and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 1996, 28, 161–219. [Google Scholar] [CrossRef]
- Loo, R.L.; Coen, M.; Ebbels, T.; Cloarec, O.; Maibaum, E.; Bictash, M.; Nicholson, J.K. Metabolic profiling and population screening of analgesic usage in nuclear magnetic resonance spectroscopy-based large-scale epidemiologic studies. Anal. Chem. 2009, 81, 5119–5129. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Loo, R.L.; Stamler, J.; Bictash, M.; Yap, I.K.; Chan, Q.; Veelkov, K.A. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008, 453, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemometr. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Tundo, G.R.; Sbardella, D.; De Pascali, S.A.; Ciaccio, C.; Coletta, M.; Fanizzi, F.P.; Marini, S. Novel Platinum(II) compounds modulate insulin-degrading enzyme activity and induce cell death in neuroblastoma cells. J. Biol. Inorg. Chem. 2015, 20, 101–108. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the tested compounds are available from the authors. |











| Metabolites | δ(1H)/ppm | Ptac2S/Controls | Cisplatin/Controls | Ptac2S/Cisplatin | |||
|---|---|---|---|---|---|---|---|
| 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | ||
| Acetate | 1.94(s) | − | − | − | − | + | |
| Alanine | 1.48(d); 3.80(m) | − | − | − | |||
| AMP | 8.23(s); 8.56(s) | − | − | − | − | ||
| Cho | 3.21(s) | + | − | + | |||
| Creatine | 3.05(s); 3.93(s) | − | − | + | − | − | + |
| Dimethylamine | 2.74(s) | − | + | − | + | − | + |
| Formate | 8.46 | − | − | ||||
| GPC | 3.24(s) | − | |||||
| Glycerol | 3.65(m) | + | + | + | + | ||
| Glycine | 3.56(s) | − | − | − | − | − | |
| Glutamine | 2.15(m); 2.45(m); 3.77(m) | − | − | − | − | − | − |
| Glutamate | 2.05(m); 2.12(m); 2.35(m) | − | − | − | − | − | − |
| Lactate | 1.32(d); 4.11(q) | − | − | − | − | + | + |
| m-inositol | 3.25(t); 3.54(dd); 3.62(t); 4.05(t) | − | − | − | |||
| Isoleucine | 0.94(t); 1.01(d); 1.98(m) | − | − | + | |||
| Leucine | 0.96(d); 0.97(d); 1.72(m); 1.73 (m) | − | + | ||||
| PC | 3.23(s) | − | − | + | − | − | + |
| Pyruvate | 2.38(s) | − | + | ||||
| Serine | 3.84(dd); 3.95(dd); 3.98(dd) | + | + | + | + | ||
| Succinate | 2.41(s) | − | − | − | − | + | |
| Taurine | 3.25(t); 3.43(t) | − | − | − | − | − | |
| TMAO | 2.9(s) | + | |||||
| UDP-Glucose | 5.97(d); 5.61(m); 6.10(d); 7.93(d) | − | − | ||||
| Valine | 1.00(d); 1.05(d); 2.28(m); 3.62(d) | − | + | ||||
| Metabolites | δ(1H)/ppm | Ptac2S/Controls | Cisplatin/Controls | Ptac2S/Cisplatin |
|---|---|---|---|---|
| Cholesterol CH3-18 | 1.94(s) | − | − | |
| Cholesterol CH3-26,27 | 1.48(d); 3.80(m) | − | − | |
| CH3 of all FAs (MUFA and DUFA) except n-3 | 8.23(s); 8.56(s) | − | − | |
| Cholesterol CH3-21 | 3.21(s) | + | ||
| all PUFAs n-3 | 3.05(s); 3.93(s) | − | + | − |
| Cholesterol CH3-19 | 2,74(s) | − | − | − |
| CH2 of all fatty chain | 8.46 | − | ||
| Cholesterol CH2 | 3.24(s) | |||
| CH2CH2COOH of all fatty acids | 3.65(m) | + | ||
| CH2CH=CH of MUFAs, DUFAs and PUFAs | 3.56(s) | − | − | − |
| CH2CH=CH of all PUFAs n-3 | 2.15(m); 2.45(m); 3.77(m) | − | − | − |
| CH2COOH of all FAs | 2.05(m); 2.12(m); 2.35(m) | − | − | − |
| CH=CH-CH2-CH=CH of DUFA and PUFA | 1.32(d); 4.11(q) | − | − | + |
| Phosphatidylcholine N-(CH3)3 | 3.25(t); 3.54(dd); 3.62 t); 4.05(t) | − | ||
| Phosphatidylcholine CH2N | 0.94(t); 1.01(d); 1.98(m) | − | − | + |
| Glycerophospholipid backbone 3-CH2 | 0.96(d); 0.97(d); 1.72(m); 1.73(m) | − | + | |
| Glycerol backbone of TG 1,3-CH2 | 3.23(s) | − | + | − |
| Phosphatidylcholine CH2OP | 2.38(s) | |||
| Glycerol backbone of TG 2-CH | 3.84(dd); 3.95(dd); 3.98(dd) | + | + | |
| CH=CH of all MUFA, DUFA, PUFA | 2.41(s) | − | − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Castro, F.; Benedetti, M.; Antonaci, G.; Del Coco, L.; De Pascali, S.A.; Muscella, A.; Marsigliante, S.; Fanizzi, F.P. Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O?-Acac)(?-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study. Molecules 2018, 23, 2301. https://doi.org/10.3390/molecules23092301
De Castro F, Benedetti M, Antonaci G, Del Coco L, De Pascali SA, Muscella A, Marsigliante S, Fanizzi FP. Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O?-Acac)(?-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study. Molecules. 2018; 23(9):2301. https://doi.org/10.3390/molecules23092301
Chicago/Turabian StyleDe Castro, Federica, Michele Benedetti, Giovanna Antonaci, Laura Del Coco, Sandra Angelica De Pascali, Antonella Muscella, Santo Marsigliante, and Francesco Paolo Fanizzi. 2018. "Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O?-Acac)(?-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study" Molecules 23, no. 9: 2301. https://doi.org/10.3390/molecules23092301
APA StyleDe Castro, F., Benedetti, M., Antonaci, G., Del Coco, L., De Pascali, S. A., Muscella, A., Marsigliante, S., & Fanizzi, F. P. (2018). Response of Cisplatin Resistant Skov-3 Cells to [Pt(O,O?-Acac)(?-Acac)(DMS)] Treatment Revealed by a Metabolomic 1H-NMR Study. Molecules, 23(9), 2301. https://doi.org/10.3390/molecules23092301








