Preparation and Identification of Antioxidative Peptides from Pacific Herring (Clupea pallasii) Protein
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cytotoxic Effects of PHPH on HepG2 Cells
2.2. Selections of Proteolytic Enzymes
2.3. Optimization of PHPH by Response Surface Methodology (RSM)
2.4. Amino Acid Composition of PHPH
2.5. Purification of PHPH
2.5.1. Fractions of PHPH by Ultrafiltration
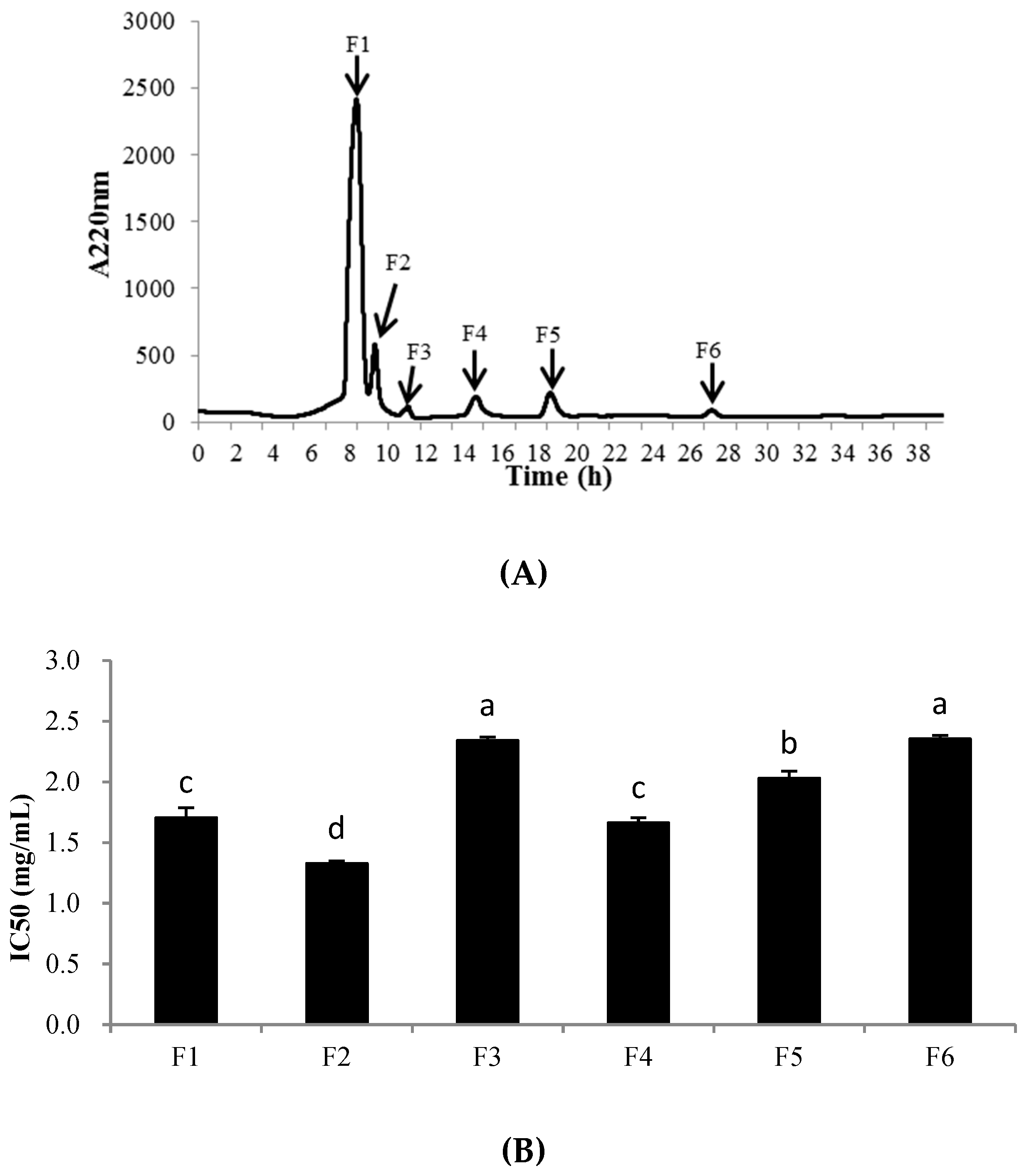
2.5.2. Gel Filtration Chromatography of PHPH-III
2.5.3. RP-HPLC Analysis
2.6. Characterization of PHPH
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Preparation of PHPH
3.3. Optimization of PHPH Preparative Conditions
3.4. Amino Acid Analysis
3.5. Antioxidant Analysis of HepG2 Cells
3.5.1. Cytotoxic Effects of PHPH on HepG2 Cells
3.5.2. Cellular Antioxidant Activity Determination
3.6. Hydroxyl Radical Scavenging Activity
3.7. DPPH Radical Scavenging Activity
3.8. IC50 Values Determination
3.9. Purification of PHPH
3.9.1. Ultrafiltration
3.9.2. Gel Filtration Chromatography
3.9.3. Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC)
3.9.4. Identification of Peptides by Mass Spectrometry
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Borawska, J.; Darewicz, M.; Pliszka, M.; Vegarud, G.E. Antioxidant properties of salmon (Salmo salar L.) protein fraction hydrolysates revealed following their ex vivo digestion and in vitro hydrolysis. J. Sci. Food Agric. 2016, 96, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.-J.; Chan, Y.-L.; Li, T.-L.; Shiau, C.-Y.; Wu, C.-J. Evaluation of lanternfish (Benthosema pterotum) hydrolysates as antioxidants against hydrogen peroxide induced oxidative injury. Food Res. Int. 2013, 54, 1409–1418. [Google Scholar] [CrossRef]
- Falowo, A.B.; Fayemi, P.O.; Muchenje, V. Natural antioxidants against lipid-protein oxidative deterioration in meat and meat products: A review. Food Res. Int. 2014, 64, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-C.; Lu, G.-H.; Jao, C.-L. Antioxidative properties of peptides prepared from tuna cooking juice hydrolysates with orientase (Bacillus subtilis). Food Res. Int. 2009, 42, 647–652. [Google Scholar] [CrossRef]
- Wolfe, K.L.; Liu, R.H. Cellular antioxidant activity (CAA) assay for assessing antioxidants, foods, and dietary supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef]
- Frankel, E.N.; Meyer, A.S. The problems of using one-dimensional methods to evaluate multifunctional food and biological antioxidants. J. Sci. Food Agric. 2000, 80, 1925–1941. [Google Scholar] [CrossRef]
- Alia, M.; Ramos, S.; Mateos, R.; Granado-Serrano, A.B.; Bravo, L.; Goya, L. Quercetin protects human hepatoma HepG2 against oxidative stress induced by tert-butyl hydroperoxide. Toxicol. Appl. Pharm. 2006, 212, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarria, B.; Mateos, R.; Gallardo, E.; Ramos, S.; Angeles Martin, M.; Bravo, L.; Goya, L. Nitroderivatives of olive oil phenols protect HepG2 cells against oxidative stress. Food Chem. Toxicol. 2012, 50, 3752–3758. [Google Scholar] [CrossRef] [Green Version]
- Yarnpakdee, S.; Benjakul, S.; Kristinsson, H.; Bakken, H. Preventive effect of Nile tilapia hydrolysate against oxidative damage of HepG2 cells and DNA mediated by H2O2 and AAPH. J. Food Sci. Technol. 2015, 52, 6194–6205. [Google Scholar] [CrossRef] [Green Version]
- Razali, N.; Mat Junit, S.; Ariffin, A.; Ramli, N.S.; Abdul Aziz, A. Polyphenols from the extract and fraction of T. indica seeds protected HepG2 cells against oxidative stress. BMC Complement. Altern. Med. 2015, 15, 438. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Bromage, E.S.; Silva, J.; Hansen, J.D.; Badil, S.M.; Woodson, J.C.; Hershberger, P.K. Production and characterization of monoclonal antibodies to IgM of Pacific herring (Clupea pallasii). Fish. Shellfish Immunol. 2012, 33, 552–558. [Google Scholar] [CrossRef]
- Hoyle, N.T.; Merritt, J.H. Quality of Fish-Protein Hydrolysates From Herring (Clupea-Harengus). J. Food Sci. 1994, 59, 76–79. [Google Scholar] [CrossRef]
- Sathivel, S.; Bechtel, P.J.; Babbitt, J.; Smiley, S.; Crapo, C.; Reppond, K.D.; Prinyawiwatkul, W. Biochemical and functional properties of herring (Clupea harengus), byproduct hydrolysates. J. Food Sci. 2003, 68, 2196–2200. [Google Scholar] [CrossRef]
- Alia, M.; Mateos, R.; Ramos, S.; Lecumberri, E.; Bravo, L.; Goya, L. Influence of quercetin and rutin on growth and antioxidant defense system of a human hepatoma cell line (HepG2). Eur. J. Nutr. 2006, 45, 19–28. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Z.; Zhao, Y.; Zeng, M. Enzymatic preparation and characterization of iron-chelating peptides from anchovy (Engraulis japonicus) muscle protein. Food Res. Int. 2012, 48, 435–441. [Google Scholar] [CrossRef]
- Ren, J.; Zhao, M.; Shi, J.; Wang, J.; Jiang, Y.; Cui, C.; Kakuda, Y.; Xue, S.J. Optimization of antioxidant peptide production from grass carp sarcoplasmic protein using response surface methodology. LWT-Food Sci. Technol. 2008, 41, 1624–1632. [Google Scholar] [CrossRef]
- Fang, X.; Xie, N.; Chen, X.; Yu, H.; Chen, J. Optimization of antioxidant hydrolysate production from flying squid muscle protein using response surface methodology. Food Bioprod. Process. 2012, 90, 676–682. [Google Scholar] [CrossRef]
- Feng, K.; Chen, W.; Sun, L.; Liu, J.; Zhao, Y.; Li, L.; Wang, Y.; Zhang, W. Optimization extraction, preliminary characterization and antioxidant activity in vitro of polysaccharides from Stachys sieboldii Miq. tubers. Carbohydr. Polym. 2015, 125, 45–52. [Google Scholar] [CrossRef]
- Shao, P.; Jiang, S.T.; Ying, Y.J. Optimization of molecular distillation for recovery of tocopherol from rapeseed oil deodorizer distillate using response surface and artificial neural network models. Food Bioprod. Process. 2007, 85, 85–92. [Google Scholar] [CrossRef]
- Batista, I.; Ramos, C.; Coutinho, J.; Bandarra, N.M.; Nunes, M.L. Characterization of protein hydrolysates and lipids obtained from black scabbardfish (Aphanopus carbo) by-products and antioxidative activity of the hydrolysates produced. Process. Biochem. 2010, 45, 18–24. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Xing, R.; Chen, X.; Liu, S.; Li, P. Optimization of antioxidative peptides from mackerel (Pneumatophorus japonicus) viscera. Peerj 2018, 6, e4373. [Google Scholar] [CrossRef]
- Sarbon, N.M.; Badii, F.; Howell, N.K. Purification and characterization of antioxidative peptides derived from chicken skin gelatin hydrolysate. Food Hydrocoll. 2018, 85, 311–320. [Google Scholar] [CrossRef]
- Sabeena Farvin, K.H.; Andersen, L.L.; Otte, J.; Nielsen, H.H.; Jessen, F.; Jacobsen, C. Antioxidant activity of cod (Gadus morhua) protein hydrolysates: Fractionation and characterisation of peptide fractions. Food Chem. 2016, 204, 409–419. [Google Scholar] [CrossRef]
- Zhang, Y.; Duan, X.; Zhuang, Y. Purification and characterization of novel antioxidant peptides from enzymatic hydrolysates of tilapia (Oreochromis niloticus) skin gelatin. Peptides 2012, 38, 13–21. [Google Scholar] [CrossRef]
- Wang, B.; Li, L.; Chi, C.F.; Ma, J.H.; Luo, H.Y.; Xu, Y.F. Purification and characterisation of a novel antioxidant peptide derived from blue mussel (Mytilus edulis) protein hydrolysate. Food Chem. 2013, 138, 1713–1719. [Google Scholar] [CrossRef]
- Ranathunga, S.; Rajapakse, N.; Kim, S.K. Purification and characterization of antioxidative peptide derived from muscle of conger eel (Conger myriaster). Eur. Food Res. Technol. 2006, 222, 310–315. [Google Scholar] [CrossRef]
- Yang, X.-R.; Zhang, L.; Ding, D.-G.; Chi, C.-F.; Wang, B.; Huo, J.-C. Preparation, Identification, and Activity Evaluation of Eight Antioxidant Peptides from Protein Hydrolysate of Hairtail (Trichiurus japonicas) Muscle. Mar. Drugs 2019, 17, 23. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A.S. Fractionation and identification of novel antioxidant peptides from fermented fish (pekasam). J. Food Meas. Charact. 2018, 12, 2174–2183. [Google Scholar] [CrossRef]
- Suetsuna, K.; Ukeda, H.; Ochi, H. Isolation and characterization of free radical scavenging activities peptides derived from casein. J. Nutr. Biochem. 2000, 11, 128–131. [Google Scholar] [CrossRef]
- Aleman, A.; Gimenez, B.; Perez-Santin, E.; Gomez-Guillen, M.C.; Montero, P. Contribution of Leu and Hyp residues to antioxidant and ACE-inhibitory activities of peptide sequences isolated from squid gelatin hydrolysate. Food Chem. 2011, 125, 334–341. [Google Scholar] [CrossRef] [Green Version]
- Umayaparvathi, S.; Arumugam, M.; Meenakshi, S.; Dräger, G.; Kirschning, A.; Balasubramanian, T. Purification and Characterization of Antioxidant Peptides from Oyster (Saccostrea cucullata) Hydrolysate and the Anticancer Activity of Hydrolysate on Human Colon Cancer Cell Lines. Int. J. Pept Res. Ther. 2013, 20, 231–243. [Google Scholar] [CrossRef]
- Kim, S.Y.; Je, J.Y.; Kim, S.K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef]
- Rajapakse, N.; Mendis, E.; Byun, H.G.; Kim, S.K. Purification and in vitro antioxidative effects of giant squid muscle peptides on free radical-mediated oxidative systems. J. Nutr. Biochem. 2005, 16, 562–569. [Google Scholar] [CrossRef]
- Chi, C.F.; Hu, F.Y.; Wang, B.; Li, Z.R.; Luo, H.Y. Influence of Amino Acid Compositions and Peptide Profiles on Antioxidant Capacities of Two Protein Hydrolysates from Skipjack Tuna (Katsuwonus pelamis) Dark Muscle. Mar. Drugs 2015, 13, 2580–2601. [Google Scholar] [CrossRef]
- Zhou, D.-Y.; Tang, Y.; Zhu, B.-W.; Qin, L.; Li, D.-M.; Yang, J.-F.; Lei, K.; Murata, Y. Antioxidant activity of hydrolysates obtained from scallop (Patinopecten yessoensis) and abalone (Haliotis discus hannai Ino) muscle. Food Chem. 2012, 132, 815–822. [Google Scholar] [CrossRef]
- Sampath Kumar, N.S.; Nazeer, R.A.; Jaiganesh, R. Purification and identification of antioxidant peptides from the skin protein hydrolysate of two marine fishes, horse mackerel (Magalaspis cordyla) and croaker (Otolithes ruber). Amino Acids 2012, 42, 1641–1649. [Google Scholar] [CrossRef]
- Uysal, S.; Cvetanovic, A.; Zengin, G.; Durovic, S.; Aktumsek, A. Optimization of the extraction process of antioxidants from loquat leaves using response surface methodology. J. Food Process. Pres. 2017, 41. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef]
- Luo, F.L.; Xing, R.E.; Wang, X.Q.; Peng, Q.C.; Li, P.C. Proximate composition, amino acid and fatty acid profiles of marine snail Rapana venosa meat, visceral mass and operculum. J. Sci. Food Agric. 2017, 97, 5361–5368. [Google Scholar] [CrossRef]
- Kong, K.W.; Mat-Junit, S.; Aminudin, N.; Hassan, F.A.; Ismail, A.; Aziz, A.A. Protective effects of the extracts of Barringtonia racemosa shoots against oxidative damage in HepG2 cells. Peerj 2016, 4. [Google Scholar] [CrossRef]
- Smeriglio, A.; Denaro, M.; Barreca, D.; Calderaro, A.; Bisignano, C.; Ginestra, G.; Bellocco, E.; Trombetta, D. In Vitro Evaluation of the Antioxidant, Cytoprotective, and Antimicrobial Properties of Essential Oil from Pistacia vera L. Variety Bronte Hull. Int. J. Mol. Sci. 2017, 18, 1212. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant Determinations By The Use Of A Stable Free Radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Luo, F.; Xing, R.; Wang, X.; Yang, H.; Li, P. Antioxidant activities of Rapana venosa meat and visceral mass during simulated gastrointestinal digestion and their membrane ultrafiltration fractions. Int. J. Food Sci. Technol. 2018, 53, 395–403. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds Pacific herring peptides are available from the authors. |


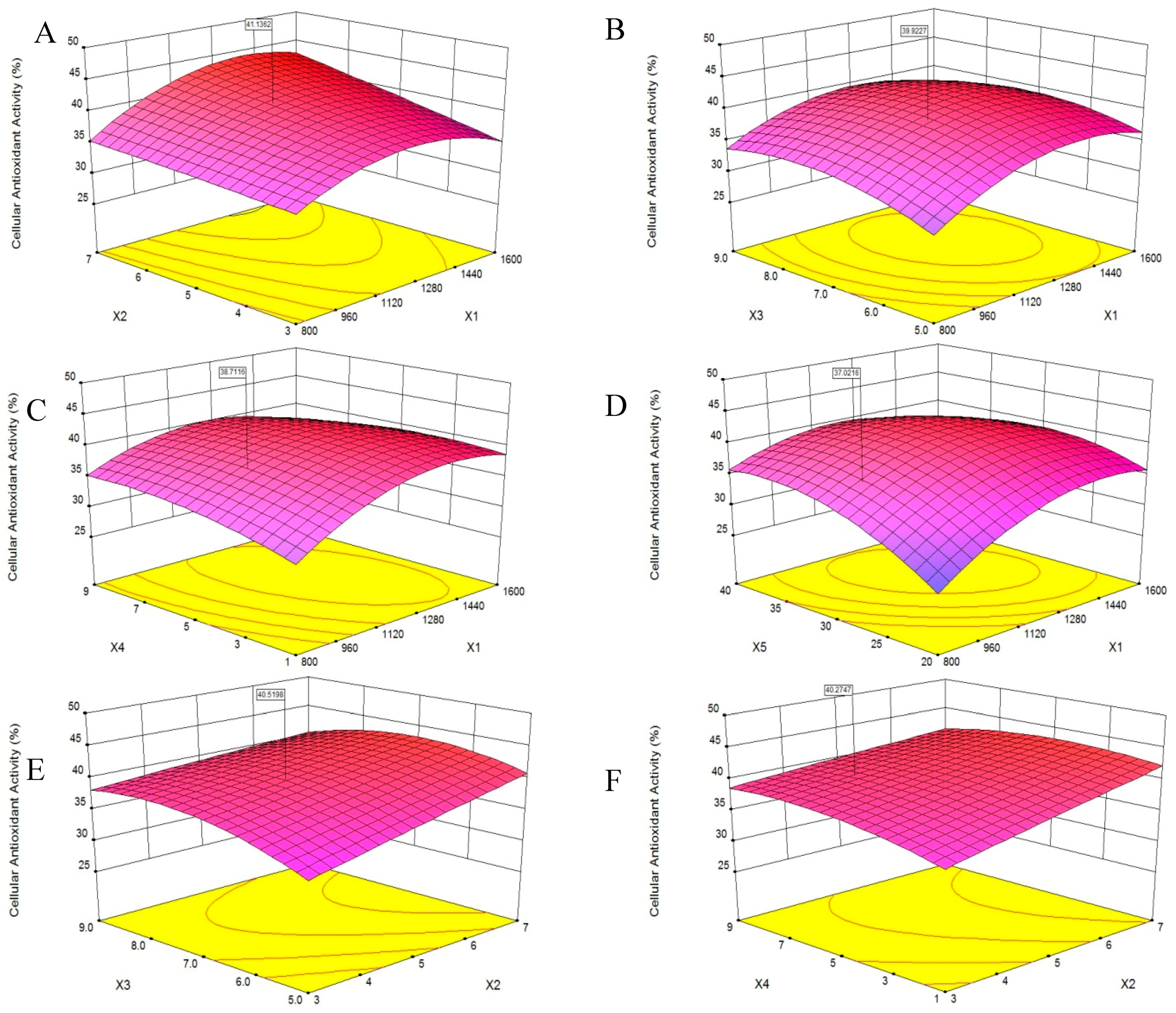





| Run Numbers | X1 | X2 | X3 | X4 | X5 | Y |
|---|---|---|---|---|---|---|
| 1 | 800 | 3 | 7 | 5 | 30 | 33.10 |
| 2 | 1600 | 3 | 7 | 5 | 30 | 34.82 |
| 3 | 800 | 7 | 7 | 5 | 30 | 36.03 |
| 4 | 1600 | 7 | 7 | 5 | 30 | 43.22 |
| 5 | 1200 | 5 | 5 | 1 | 30 | 36.64 |
| 6 | 1200 | 5 | 9 | 1 | 30 | 38.06 |
| 7 | 1200 | 5 | 5 | 9 | 30 | 36.34 |
| 8 | 1200 | 5 | 9 | 9 | 30 | 37.15 |
| 9 | 1200 | 3 | 7 | 5 | 20 | 32.29 |
| 10 | 1200 | 7 | 7 | 5 | 20 | 36.03 |
| 11 | 1200 | 3 | 7 | 5 | 40 | 37.60 |
| 12 | 1200 | 7 | 7 | 5 | 40 | 42.72 |
| 13 | 800 | 5 | 5 | 5 | 30 | 29.23 |
| 14 | 1600 | 5 | 5 | 5 | 30 | 36.81 |
| 15 | 800 | 5 | 9 | 5 | 30 | 33.86 |
| 16 | 1600 | 5 | 9 | 5 | 30 | 38.19 |
| 17 | 1200 | 5 | 7 | 1 | 20 | 34.06 |
| 18 | 1200 | 5 | 7 | 9 | 20 | 35.43 |
| 19 | 1200 | 5 | 7 | 1 | 40 | 35.63 |
| 20 | 1200 | 5 | 7 | 9 | 40 | 35.73 |
| 21 | 1200 | 3 | 5 | 5 | 30 | 32.55 |
| 22 | 1200 | 7 | 5 | 5 | 30 | 37.85 |
| 23 | 1200 | 3 | 9 | 5 | 30 | 37.93 |
| 24 | 1200 | 7 | 9 | 5 | 30 | 40.54 |
| 25 | 800 | 5 | 7 | 1 | 30 | 31.42 |
| 26 | 1600 | 5 | 7 | 1 | 30 | 34.81 |
| 27 | 800 | 5 | 7 | 9 | 30 | 31.34 |
| 28 | 1600 | 5 | 7 | 9 | 30 | 34.55 |
| 29 | 1200 | 5 | 5 | 5 | 20 | 31.51 |
| 30 | 1200 | 5 | 9 | 5 | 20 | 32.90 |
| 31 | 1200 | 5 | 5 | 5 | 40 | 33.42 |
| 32 | 1200 | 5 | 9 | 5 | 40 | 37.82 |
| 33 | 800 | 5 | 7 | 5 | 20 | 25.73 |
| 34 | 1600 | 5 | 7 | 5 | 20 | 30.13 |
| 35 | 800 | 5 | 7 | 5 | 40 | 36.72 |
| 36 | 1600 | 5 | 7 | 5 | 40 | 40.38 |
| 37 | 1200 | 3 | 7 | 1 | 30 | 34.52 |
| 38 | 1200 | 7 | 7 | 1 | 30 | 41.03 |
| 39 | 1200 | 3 | 7 | 9 | 30 | 42.03 |
| 40 | 1200 | 7 | 7 | 9 | 30 | 43.22 |
| 41 | 1200 | 5 | 7 | 5 | 30 | 39.07 |
| 42 | 1200 | 5 | 7 | 5 | 30 | 38.99 |
| 43 | 1200 | 5 | 7 | 5 | 30 | 42.80 |
| 44 | 1200 | 5 | 7 | 5 | 30 | 39.49 |
| 45 | 1200 | 5 | 7 | 5 | 30 | 42.14 |
| 46 | 1200 | 5 | 7 | 5 | 30 | 41.14 |
| Variables | Sum of Squares | DF | Mean Square | F Value | P Value |
|---|---|---|---|---|---|
| Model | 601.75 | 20 | 30.09 | 15.34 | <0.0001 |
| Residual | 49.05 | 25 | 1.96 | ||
| Lack of fit | 38.51 | 20 | 1.93 | 0.91 | 0.6059 |
| Pure error | 10.53 | 5 | 2.11 | ||
| Cor total | 650.80 | 45 | |||
| R2 | 0.9246 | ||||
| Adj. R2 | 0.8643 | ||||
| Pred. R2 | 0.7400 | ||||
| Adeq precision | 17.822 | ||||
| CV% | 3.80 |
| Amino Acid | Content (%) |
|---|---|
| Asp | 4.93 |
| Thr * | 2.26 |
| Ser | 2.19 |
| Glu | 6.22 |
| Gly | 3.5 |
| Ala | 2.78 |
| Gys | 0.11 |
| Val * | 2.95 |
| Met * | 1.14 |
| Ile * | 2.45 |
| Leu * | 4.43 |
| Tyr | 1.07 |
| Phe * | 2.43 |
| Lys * | 4.3 |
| His | 1.35 |
| Arg | 4.01 |
| Pro | 2.37 |
| ∑AA | 48.49 |
| ∑EAA | 19.96 |
| ∑EAA/∑NEAA | 0.70 |
| ∑EAA/∑AA | 0.41 |
| Sample | IC50 (mg/mL) | ||
|---|---|---|---|
| Hydroxyl Radical Scavenging Activity | DPPH Radical Scavenging Activity | Cellular Antioxidant Activity | |
| P1 | 4.57 ± 0.24 | 5.14 ± 0.32 | 1.19 ± 0.05 |
| P2 | 3.78 ± 0.17 | 4.37 ± 0.26 | 1.04 ± 0.06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yu, H.; Xing, R.; Liu, S.; Chen, X.; Li, P. Preparation and Identification of Antioxidative Peptides from Pacific Herring (Clupea pallasii) Protein. Molecules 2019, 24, 1946. https://doi.org/10.3390/molecules24101946
Wang X, Yu H, Xing R, Liu S, Chen X, Li P. Preparation and Identification of Antioxidative Peptides from Pacific Herring (Clupea pallasii) Protein. Molecules. 2019; 24(10):1946. https://doi.org/10.3390/molecules24101946
Chicago/Turabian StyleWang, Xueqin, Huahua Yu, Ronge Xing, Song Liu, Xiaolin Chen, and Pengcheng Li. 2019. "Preparation and Identification of Antioxidative Peptides from Pacific Herring (Clupea pallasii) Protein" Molecules 24, no. 10: 1946. https://doi.org/10.3390/molecules24101946