Novel Derivatives of Deoxycholic Acid Bearing Linear Aliphatic Diamine and Aminoalcohol Moieties and their Cyclic Analogs at the C3 Position: Synthesis and Evaluation of Their In Vitro Antitumor Potential
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Biological Evaluation
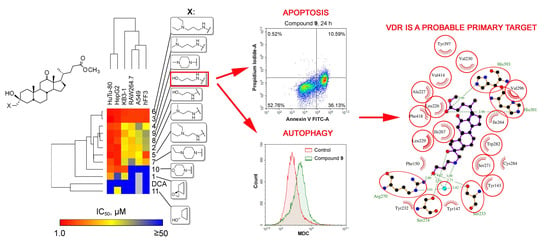
2.2.1. Cytotoxicity of Novel DCA Derivatives and SAR Analysis
- (a)
- Replacement of terminal tertiary amino groups in the side moiety by the hydroxyl group is highly promising—derivatives 8 and 9, containing aminoalcohol substituents, were found to display more pronounced selectivity for tested tumor cells in comparison with compounds, bearing corresponding diamine-containing group (cf., 8 to 2/5 and 9 to 3/6);
- (b)
- Elongation of the hydrocarbon chain between hydroxyl and amino groups has almost no effect on cytotoxicity (IC508 = 4.4–34.8 µM, IC509 = 4.0–33.3 µM), whereas, in the case of diamine-bearing derivatives, it was shown that a similar modification significantly increased the activity (IC502 = 7.1–36.0 µM and IC503 = 3.0–6.8 µM; IC505 = 3.3–25.5 µM and IC506 = 1.0–6.8 µM).
- (c)
- Replacement of a hydroxyl group by an ether group, which limits the conformational flexibility, markedly reduces the cytotoxicity of derivatives with respect to non-malignant fibroblasts hFF3. Thus, compound 10, bearing a morpholine substituent, is characterized by the highest IC50 values for this cell line (IC50hFF3 > 50 µM) among all tested DCA derivatives. However, this modification decreased the water solubility of the derivative and, therefore, limited its consideration as a lead compound for further studies.
2.2.2. Inhibitory Activity of Novel DCA Derivatives against NO Synthesis by Macrophages
2.2.3. In silico ADME Analysis
2.2.4. Mechanism of Cytotoxic Effect of Compound 9
Compound 9 Induces Intrinsic Caspase-Dependent Apoptosis
Compound 9 Triggers Cytodestructive Autophagy
VDR is a Probable Primary Target of Compound 9
3. Materials and Methods
3.1. General Experimental Procedures and Reagents
3.2. General Procedure for the Synthesis of Compounds 8–10
3.2.1. Methyl 3-hydroxy-3-((2-hydroxyethylamino)methyl)-12-oxo-5β-cholan-24-oate (8)
3.2.2. Methyl 3-hydroxy-3-((3-hydroxypropylamino)methyl)-12-oxo-5β-cholan-24-oate (9)
3.2.3. Methyl 3-hydroxy-3-(morpholinomethyl)-12-oxo-5β-cholan-24-oate (10)
3.3. Cell Cultures and DCA Derivatives
3.4. Evaluation of Cytotoxicity of DCA Derivatives by MTT Assay
3.5. Inhibition of NO Production by Stimulated Macrophages
3.6. In Silico ADME Prediction
3.7. Annexin V FITC/PI Apoptosis Detection
3.8. Cell Cycle Assay
3.9. Determination of Mitochondrial Membrane Potential (∆ψM)
3.10. Determination ofCcaspase-3/-7 Activity
3.11. Autophagy Detection
3.12. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofmann, A.F.; Hagey, L.R. Key discoveries in bile acid chemistry and biology and their clinical applications: History of the last eight decades. J. Lipid Res. 2014, 55, 1553–1595. [Google Scholar] [CrossRef] [PubMed]
- Arlia-Ciommo, A.; Piano, A.; Svistkova, V.; Mohtashami, S.; Titorenko, V.I. Mechanisms underlying the anti-aging and anti-tumor effects of lithocholic bile acid. Int. J. Mol. Sci. 2014, 15, 16522–16543. [Google Scholar] [CrossRef] [PubMed]
- Popadyuk, I.I.; Salomatina, O.V.; Salakhutdinov, N.F. Modern approaches to modification of bile acids for the synthesis of compounds possessing valuable physicochemical and biological properties. Russ. Chem. Rev. 2017, 86, 388–443. [Google Scholar] [CrossRef]
- Sharma, R.; Long, A.; Gilmer, J.F. Advances in bile acid medicinal chemistry. Curr. Med. Chem. 2011, 18, 4029–4052. [Google Scholar] [CrossRef]
- Salvador, J.A.R.; Carvalho, J.F.S.; Neves, M.A.C.; Silvestre, S.M.; Leitão, A.J.; Silva, M.M.C.; Sá, E.; Melo, M.L. Anticancer Steroids: Linking Natural and Semi-Synthetic Compounds. Nat. Prod. Rep. 2013, 30, 324–374. [Google Scholar] [CrossRef]
- Michalson, E.T.; Szmuszkovicz, J. Medicinal agents incorporating the 1,2-diamine functionality. In Progress in Drug Research; Jucker, E., Ed.; Birkhäuser Basel: Basel, Switzerland, 1989; pp. 135–149. [Google Scholar]
- Levitt, R.C.; Nicolaides, N.C.; Kinney, W.A.; Jones, S. Asthma Associated Factors as Targets for Treating Atopic Allergies Including Asthma and Related Disorders. U.S. Patent US5908839A, 1 June 1999. [Google Scholar]
- El Kihel, L.; Clément, M.; Bazin, M.A.; Descamps, G.; Khalid, M.; Rault, S. New lithocholic and chenodeoxycholic piperazinylcarboxamides with antiproliferative and pro-apoptotic effects on human cancer cell lines. Bioorgan. Med. Chem. 2008, 16, 8737–8744. [Google Scholar] [CrossRef]
- Salunke, D.B.; Hazra, B.G.; Pore, V.S.; Bhat, M.K.; Nahar, P.B.; Deshpande, M.V. New Steroidal Dimers with Antifungal and Antiproliferative Activity. J. Med. Chem. 2004, 47, 1591–1594. [Google Scholar] [CrossRef]
- McLane, M.; Ruiz-White, I.; Maloy, W.L.; Wolfe, H.R. Aminosteroids for the Treatment of a PTP1B Associated Disease. U.S. Patent US9546194B2, 17 January 2017. [Google Scholar]
- Mclane, M.; Ruiz-White, I.; Wolfe, H. A Method for Treating Diabetes. International Patent WO2009032321A3, 12 March 2009. [Google Scholar]
- Ayan, D.; Maltais, R.; Poirier, D. Identification of a 17β-Hydroxysteroid Dehydrogenase Type 10 Steroidal Inhibitor: A Tool to Investigate the Role of Type 10 in Alzheimer’s Disease and Prostate Cancer. ChemMedChem 2012, 7, 1181–1184. [Google Scholar] [CrossRef]
- Maltais, R.; Fournier, M.A.; Poirier, D. Development of 3-substituted-androsterone derivatives as potent inhibitors of 17β-hydroxysteroid dehydrogenase type 3. Bioorgan. Med. Chem. 2011, 19, 4652–4668. [Google Scholar] [CrossRef]
- Vicens, M.; Medarde, M.; Macias, R.I.R.; Larena, M.G.; Villafaina, A.; Serrano, M.A.; Marin, J.J.G. Novel cationic and neutral glycocholic acid and polyamine conjugates able to inhibit transporters involved in hepatic and intestinal bile acid uptake. Bioorgan. Med. Chem. 2007, 15, 2359–2367. [Google Scholar] [CrossRef]
- Vicens, M.; Macias, R.I.R.; Briz, O.; Rodriguez, A.; El-Mir, M.Y.; Medarde, M.; Marin, J.J.G. Inhibition of the intestinal absorption of bile acids using cationic derivatives: Mechanism and repercussions. Biochem. Pharmacol. 2007, 73, 394–404. [Google Scholar] [CrossRef]
- Bülbül, M.; Saraçoğlu, N.; Küfrevioğlu, O.I.; Ciftçi, M. Bile acid derivatives of 5-amino-1,3,4-thiadiazole-2-sulfonamide as new carbonic anhydrase inhibitors: Synthesis and investigation of inhibition effects. Bioorgan. Med. Chem. 2002, 10, 2561–2567. [Google Scholar] [CrossRef]
- Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors. Preparation of potent sulfonamides inhibitors incorporating bile acid tails. Bioorgan. Med. Chem. Lett. 2002, 12, 1551–1557. [Google Scholar] [CrossRef]
- Hazra, B.G.; Pore, V.S.; Dey, S.K.; Datta, S.; Darokar, M.P.; Saikia, D.; Khanuja, S.P.S.; Thakur, A.P. Bile acid amides derived from chiral amino alcohols: Novel antimicrobials and antifungals. Bioorgan. Med. Chem. Lett. 2004, 14, 773–777. [Google Scholar] [CrossRef]
- Luo, G.; Qian, Z.; Qiu, R.; You, Q.; Xiang, H. Lipid reducing activity of novel cholic acid (CA) analogs: Design, synthesis and preliminary mechanism study. Bioorgan. Chem. 2018, 80, 396–407. [Google Scholar] [CrossRef]
- Valkonen, A.; Lahtinen, M.; Virtanen, E.; Kaikkonen, S.; Kolehmainen, E. Bile acid amidoalcohols: Simple organogelators. Biosens. Bioelectron. 2004, 20, 1233–1241. [Google Scholar] [CrossRef]
- Popadyuk, I.I.; Markov, A.V.; Babich, V.O.; Salomatina, O.V.; Logashenko, E.B.; Zenkova, M.A.; Salakhutdinov, N.F. Novel derivatives of deoxycholic acid bearing aliphatic or cyclic diamine moieties at the C-3 position: Synthesis and evaluation of anti-proliferative activity. Bioorgan. Med. Chem. Lett. 2017, 27, 3755–3759. [Google Scholar] [CrossRef]
- Popadyuk, I.I.; Markov, A.V.; Salomatina, O.V.; Logashenko, E.B.; Shernyukov, A.V.; Zenkova, M.A.; Salakhutdinov, N.F. Synthesis and biological activity of novel deoxycholic acid derivatives. Bioorgan. Med. Chem. 2015, 23, 5022–5034. [Google Scholar] [CrossRef]
- Popadyuk, I.I.; Markov, A.V.; Morozova, E.A.; Babich, V.O.; Salomatina, O.V.; Logashenko, E.B.; Zenkova, M.A.; Tolstikova, T.G.; Salakhutdinov, N.F. Synthesis and evaluation of antitumor, anti-inflammatory and analgesic activity of novel deoxycholic acid derivatives bearing aryl- or hetarylsulfanyl moieties at the C-3 position. Steroids 2017, 127, 1–12. [Google Scholar] [CrossRef]
- Deuk Kim, N.; Im, E.; Hyun Yoo, Y.; Hyun Choi, Y. Modulation of the Cell Cycle and Induction of Apoptosis in Human Cancer Cells by Synthetic Bile Acids. Curr. Cancer Drug Targets 2006, 6, 681–689. [Google Scholar] [CrossRef]
- Blanchet, M.; Brunel, J.M. Bile Acid Derivatives: From Old Molecules to a New Potent Therapeutic Use: An Overview. Curr. Med. Chem. 2018, 25, 3613–3636. [Google Scholar] [CrossRef]
- Hofmann, A.F. The enterohepatic circulation of bile acids in mammals: Form and functions. Front. Biosci. 2009, 14, 2584–2598. [Google Scholar] [CrossRef]
- Fukumura, D.; Kashiwagi, S.; Jain, R.K. The role of nitric oxide in tumour progression. Nat. Rev. Cancer 2006, 6, 521–534. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Mariño, G.; Kroemer, G. Mechanisms of apoptotic phosphatidylserine exposure. Cell Res. 2013, 23, 1247–1248. [Google Scholar] [CrossRef]
- Smiley, S.T.; Reers, M.; Mottola-Hartshorn, C.; Lin, M.; Chen, A.; Smith, T.W.; Steele, G.D.; Chen, L.B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. USA 1991, 88, 3671–3675. [Google Scholar] [CrossRef]
- Roesly, H.B.; Khan, M.R.; Chen, H.D.R.; Hill, K.A.; Narendran, N.; Watts, G.S.; Chen, X.; Dvorak, K. The decreased expression of Beclin-1 correlates with progression to esophageal adenocarcinoma: The role of deoxycholic acid. Am. J. Physiol. Liver Physiol. 2012, 302, G864–G872. [Google Scholar] [CrossRef]
- Gafar, A.A.; Draz, H.M.; Goldberg, A.A.; Bashandy, M.A.; Bakry, S.; Khalifa, M.A.; AbuShair, W.; Titorenko, V.I.; Sanderson, J.T. Lithocholic acid induces endoplasmic reticulum stress, autophagy and mitochondrial dysfunction in human prostate cancer cells. PeerJ 2016, 4, e2445. [Google Scholar] [CrossRef]
- Eskelinen, E.L.; Saftig, P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 664–673. [Google Scholar] [CrossRef]
- Clarke, P.G.H. Developmental cell death: Morphological diversity and multiple mechanisms. Anat. Embryol. 1990, 181, 195–213. [Google Scholar] [CrossRef]
- Contento, A.L.; Xiong, Y.; Bassham, D.C. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005, 42, 598–608. [Google Scholar] [CrossRef]
- Simon, H.U.; Haj-Yehia, A.; Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis 2000, 5, 415–418. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Elazar, Z. Regulation of autophagy by ROS: Physiology and pathology. Trends Biochem. Sci. 2011, 36, 30–38. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Zhou, H.; Pandak, W.M.; Ren, S.; Gil, G.; Dent, P. Bile acids as regulatory molecules. J. Lipid Res. 2009, 50, 1509–1520. [Google Scholar] [CrossRef]
- Schaap, F.G.; Trauner, M.; Jansen, P.L.M. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 55–67. [Google Scholar] [CrossRef]
- Ishizawa, M.; Matsunawa, M.; Adachi, R.; Uno, S.; Ikeda, K.; Masuno, H.; Shimizu, M.; Iwasaki, K.; Yamada, S.; Makishima, M. Lithocholic acid derivatives act as selective vitamin D receptor modulators without inducing hypercalcemia. J. Lipid Res. 2008, 49, 763–772. [Google Scholar] [CrossRef]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef]
- Høyer-Hansen, M.; Nordbrandt, S.P.S.; Jäättelä, M. Autophagy as a basis for the health-promoting effects of vitamin D. Trends Mol. Med. 2010, 16, 295–302. [Google Scholar] [CrossRef]
- Tavera-Mendoza, L.E.; Westerling, T.; Libby, E.; Marusyk, A.; Cato, L.; Cassani, R.; Cameron, L.A.; Ficarro, S.B.; Marto, J.A.; Klawitter, J.; et al. Vitamin D receptor regulates autophagy in the normal mammary gland and in luminal breast cancer cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2186–E2194. [Google Scholar] [CrossRef]
- Kizildag, S.; Ates, H.; Kizildag, S. Treatment of K562 cells with 1,25-dihydroxyvitamin D3 induces distinct alterations in the expression of apoptosis-related genes BCL2, BAX, BCLXL, and p21. Ann. Hematol. 2010, 89, 1–7. [Google Scholar] [CrossRef]
- Guzey, M.; Kitada, S.; Reed, J.C. Apoptosis induction by 1alpha,25-dihydroxyvitamin D3 in prostate cancer. Mol. Cancer Ther. 2002, 1, 667–677. [Google Scholar]
- Ferronato, M.J.; Alonso, E.N.; Gandini, N.A.; Fermento, M.E.; Villegas, M.E.; Quevedo, M.A.; Arévalo, J.; López Romero, A.; Rivadulla, M.L.; Gómez, G.; et al. The UVB1 Vitamin D analogue inhibits colorectal carcinoma progression. J. Steroid Biochem. Mol. Biol. 2016, 163, 193–205. [Google Scholar] [CrossRef]
- Wang, C.; Wang, B.; Hou, S.; Xue, L.; Kang, Z.; Du, J.; Li, Y.; Liu, X.; Wang, Q.; Zhang, C. Discovery of novel nonsteroidal VDR agonists with novel diarylmethane skeleton for the treatment of breast cancer. Eur. J. Med. Chem. 2019, 163, 787–803. [Google Scholar] [CrossRef]
- Abu el Maaty, M.A.; Strassburger, W.; Qaiser, T.; Dabiri, Y.; Wölfl, S. Differences in p53 status significantly influence the cellular response and cell survival to 1,25-dihydroxyvitamin D3-metformin cotreatment in colorectal cancer cells. Mol. Carcinog. 2017, 56, 2486–2498. [Google Scholar] [CrossRef]
- Wang, R.C.; Levine, B. Calcipotriol Induces Autophagy in HeLa Cells and Keratinocytes. J. Invest. Dermatol. 2011, 131, 990–993. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |





| Compound | IC50 (µM) | IC50NO (µM) | |||||
|---|---|---|---|---|---|---|---|
| Duodenum | Liver | Cervix | Lung | Macrophage | Fibroblast | ||
| HuTu-80 | HepG2 | KB3-1 | A549 | RAW264.7 | hFF3 | ||
| 1 * | 17.0 ± 1.6 * | 43.5 ± 1.9 * | 27.9 ± 2.5 * | ND | >100 | 53.9 ± 3.3 * | 16.4 ± 0.9 |
| 2 * | 7.1 ± 0.6 * | 8.0 ± 0.9 * | 26.2 ± 0.8 * | 36 ± 0.7 * | 21.7 ± 0.87 | 25.4 ± 2.2 * | >10 |
| 3 * | 3.0 ± 0.4 * | 4.0 ± 0.2 * | 5.9 ± 0.5 * | 6.8 ± 0.1 * | 6.7 ± 0.4 | 6.5 ± 0.5 * | >10 |
| 4 * | 4.7 ± 0.2 * | 3.6 ± 0.7 * | 23.5 ± 1.7 * | 21.6 ± 2.5 * | 20.4 ± 0.4 | 32.5 ± 1.4 * | >10 |
| 5 * | 3.3 ± 0.1 * | 4.3 ± 0.3 * | 21.6 ± 3.4 * | 25.5 ± 0.8 * | 20.9 ± 2.1 | 7.4 ± 1.0 * | >10 |
| 6 * | 1.0 ± 0.6 * | 3.7 ± 0.17 * | 6.7 ± 0.6 * | 6.8 ± 0.3 * | 6.7 ± 0.5 | 6.2 ± 0.4 * | >10 |
| 7 * | 8.3 ± 1.2 * | 3.8 ± 0.3 * | 28.6 ± 0.5 * | 23.5 ± 3.0 * | 37.8 ± 1.7 | 17.4 ± 1.6 * | >10 |
| 8 | 4.4 ± 0.34 | 5.8 ± 1.3 | 17.7 ± 2.8 | 29.6 ± 2.5 | 23.4 ± 1.2 | 34.8 ± 1.0 | >10 |
| 9 | 4.0 ± 0.5 | 7.3 ± 0.6 | 22.8 ± 1.8 | 28.98 ± 3.5 | 19.3 ± 3.7 | 33.3 ± 1.2 | >10 |
| 10 | 10 ± 0.3 | 7.5 ± 1.2 | 8.4 ± 0.5 | >50 | >50 | >50 | >10 |
| 11 * | >100 * | 25.7 ± 3.6 * | >100 * | ND | >100 | >100 * | 50.9 ± 5.9 |
| DCA * | 82.9 ± 1.9 * | >100 * | >100 * | ND | >100 | >100 * | 83.6 ± 1.6 |
| Compound | MW (Da) a | mLogP b | HBA c | HBD d | Lipinski Violation |
|---|---|---|---|---|---|
| Rule | ≤500 | ≤5 | ≤10 | ≤5 | 0 |
| 1 | 477.68 | 2.94 | 6 | 3 | 0 |
| 2 | 504.74 | 3.33 | 6 | 2 | 1 |
| 3 | 518.77 | 3.52 | 6 | 2 | 1 |
| 4 | 516.76 | 3.52 | 6 | 1 | 1 |
| 5 | 532.8 | 3.71 | 6 | 2 | 1 |
| 6 | 546.82 | 3.89 | 6 | 2 | 1 |
| 7 | 530.78 | 3.71 | 6 | 1 | 1 |
| 8* | 477.68 | 2.94 | 6 | 3 | 0 |
| 9* | 491.7 | 3.14 | 6 | 3 | 0 |
| 10 | 503.71 | 3.33 | 6 | 1 | 1 |
| 11 | 404.58 | 3.98 | 4 | 1 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markov, A.V.; Babich, V.O.; Popadyuk, I.I.; Salomatina, O.V.; Logashenko, E.B.; Salakhutdinov, N.F.; Zenkova, M.A. Novel Derivatives of Deoxycholic Acid Bearing Linear Aliphatic Diamine and Aminoalcohol Moieties and their Cyclic Analogs at the C3 Position: Synthesis and Evaluation of Their In Vitro Antitumor Potential. Molecules 2019, 24, 2644. https://doi.org/10.3390/molecules24142644
Markov AV, Babich VO, Popadyuk II, Salomatina OV, Logashenko EB, Salakhutdinov NF, Zenkova MA. Novel Derivatives of Deoxycholic Acid Bearing Linear Aliphatic Diamine and Aminoalcohol Moieties and their Cyclic Analogs at the C3 Position: Synthesis and Evaluation of Their In Vitro Antitumor Potential. Molecules. 2019; 24(14):2644. https://doi.org/10.3390/molecules24142644
Chicago/Turabian StyleMarkov, Andrey V., Valeriya O. Babich, Irina I. Popadyuk, Oksana V. Salomatina, Evgeniya B. Logashenko, Nariman F. Salakhutdinov, and Marina A. Zenkova. 2019. "Novel Derivatives of Deoxycholic Acid Bearing Linear Aliphatic Diamine and Aminoalcohol Moieties and their Cyclic Analogs at the C3 Position: Synthesis and Evaluation of Their In Vitro Antitumor Potential" Molecules 24, no. 14: 2644. https://doi.org/10.3390/molecules24142644
APA StyleMarkov, A. V., Babich, V. O., Popadyuk, I. I., Salomatina, O. V., Logashenko, E. B., Salakhutdinov, N. F., & Zenkova, M. A. (2019). Novel Derivatives of Deoxycholic Acid Bearing Linear Aliphatic Diamine and Aminoalcohol Moieties and their Cyclic Analogs at the C3 Position: Synthesis and Evaluation of Their In Vitro Antitumor Potential. Molecules, 24(14), 2644. https://doi.org/10.3390/molecules24142644