Factors Influencing the Activity of Nanozymes in the Cleavage of an RNA Model Substrate
Abstract
:1. Introduction
2. Results and Discussion
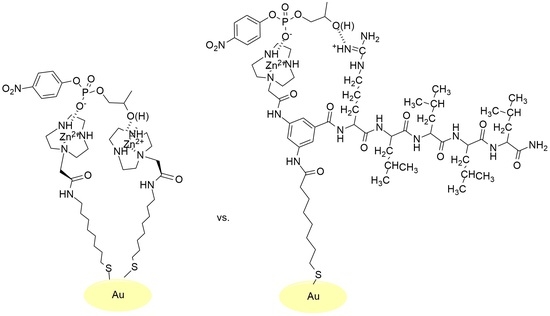
2.1. Synthesis of the Thiols and Passivation of AuNPs
2.2. Kinetics of the Cleavage of HPNP in the Presence of the Zn(II) Complexes of AuNP1-5.
2.2.1. Cooperativity between Functional Groups
2.2.2. Michaelis–Menten Kinetics
2.3. The Source of Catalysis in Zn(II)-Triazacyclononane-Functionalized Nanozymes
2.3.1. Dinuclear Catalysts AuNP1-3
2.3.2. Mononuclear Catalysts AuNP4 and AuNP5
3. Materials and Methods
3.1. General
3.2. Preparation of the AuNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palermo, G.; Cavalli, A.; Klein, M.; Alfonso-Prieto, M.; Peraro, M.; Vivo, M. Catalytic Metal Ions and Enzymatic Processing of DNA and RNA. Accounts Chem. Res. 2015, 48, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Mancin, F.; Scrimin, P.; Tecilla, P. Progress in artificial metallonucleases. Chem. Commun. 2012, 48, 5545–5559. [Google Scholar] [CrossRef] [PubMed]
- Diez-Castellnou, M.; Martinez, A.; Mancin, F. Phosphate Ester Hydrolysis: The Path From Mechanistic Investigation to the Realization of Artificial Enzymes. Ad. Phys. Org. Chem. 2017, 51, 129–186. [Google Scholar]
- Wolfenden, R. Benchmark Reaction Rates, the Stability of Biological Molecules in Water, and the Evolution of Catalytic Power in Enzymes. Annu. Rev. Biochem. 2011, 80, 645–667. [Google Scholar] [CrossRef] [PubMed]
- Yatsimirsky, A. Metal ion catalysis in acyl and phosphoryl transfer: Transition states as ligands. Coordin Chem. Rev. 2005, 249, 1997–2011. [Google Scholar] [CrossRef]
- Tirel, E.; Bellamy, Z.; Adams, H.; Lebrun, V.; Duarte, F.; Williams, N. Catalytic Zinc Complexes for Phosphate Diester Hydrolysis. Angew. Chem. Int. Ed. 2014, 53, 8246–8250. [Google Scholar] [CrossRef] [Green Version]
- Dupureur, C. Roles of metal ions in nucleases. Curr. Opin. Chem. Biol. 2008, 12, 250–255. [Google Scholar] [CrossRef]
- Manea, F.; Houillon, F.B.; Pasquato, L.; Scrimin, P. Nanozymes: Gold-nanoparticle-based transphosphorylation catalysts. Angew. Chem. Int. Ed. Engl. 2004, 43, 6165–6169. [Google Scholar] [CrossRef]
- Mancin, F.; Prins, L.; Pengo, P.; Pasquato, L.; Tecilla, P.; Scrimin, P. Hydrolytic Metallo-Nanozymes: From Micelles and Vesicles to Gold Nanoparticles. Molecules 2016, 21, 1014. [Google Scholar] [CrossRef]
- Bonomi, R.; Selvestrel, F.; Lombardo, V.; Sissi, C.; Polizzi, S.; Mancin, F.; Tonellato, U.; Scrimin, P. Phosphate Diester and DNA Hydrolysis by a Multivalent, Nanoparticle-Based Catalyst. J. Am. Chem. Soc. 2008, 130, 15744–15745. [Google Scholar] [CrossRef]
- Martin, M.; Manea, F.; Fiammengo, R.; Prins, L.J.; Pasquato, L.; Scrimin, P. Metallodendrimers as transphosphorylation catalysts. J. Am. Chem. Soc. 2007, 129, 6982–6983. [Google Scholar] [CrossRef]
- Knight, A.; Nita, R.; Moore, M.; Zabetakis, D.; Khandelwal, M.; Martin, B.; Fontana, J.; Goldberg, E.; Funk, A.; Chang, E.; et al. Surface plasmon resonance promotion of homogeneous catalysis using a gold nanoparticle platform. J. Nanopart Res. 2014, 16, 2400. [Google Scholar] [CrossRef]
- Khulbe, K.; Roy, P.; Radhakrishnan, A.; Mugesh, G. An Unusual Two-Step Hydrolysis of Nerve Agents by a Nanozyme. Chemcatchem 2018, 10, 4826–4831. [Google Scholar] [CrossRef]
- Zhang, Z.; Fu, Q.; Li, X.; Huang, X.; Xu, J.; Shen, J.; Liu, J. Self-assembled gold nanocrystal micelles act as an excellent artificial nanozyme with ribonuclease activity. J. Biol. Inorg. Chem. 2009, 14, 653–662. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, X.; Fong, L.; Margerum, L. Ligand effects on the phosphoesterase activity of Co(II) Schiff base complexes built on PAMAM dendrimers. Inorg. Chim. Acta 2001, 317, 72–80. [Google Scholar] [CrossRef]
- Bazzicalupi, C.; Bianchi, A.; Giorgi, C.; Valtancoli, B. Zn(II) enhances nucleotide binding and dephosphorylation in the presence of a poly(ethylene imine) dendrimer. Inorg. Chim. Acta 2014, 417, 163–170. [Google Scholar] [CrossRef]
- Bonomi, R.; Scrimin, P.; Mancin, F. Phosphate diesters cleavage mediated by Ce(IV) complexes self-assembled on gold nanoparticles. Org. Biomol. Chem. 2010, 8, 2622–2626. [Google Scholar] [CrossRef]
- Williams, N.H.; Takasaki, B.; Wall, M.; Chin, J. Structure and Nuclease Activity of Simple Dinuclear Metal Complexes: Quantitative Dissection of the Role of Metal Ions. Acc. Chem. Res. 1999, 32, 485–493. [Google Scholar] [CrossRef]
- Brown, R.S. Bio-inspired approaches to accelerating metal ion-promoted reactions: Enzyme-like rates for metal ion mediated phosphoryl and acyl transfer processes. Pure Appl. Chem. 2015, 87, 601–614. [Google Scholar] [CrossRef]
- Diez-Castellnou, M.; Mancin, F.; Scrimin, P. Efficient phosphodiester cleaving nanozymes resulting from multivalency and local medium polarity control. J. Am. Chem. Soc. 2014, 136, 1158–1161. [Google Scholar] [CrossRef]
- Scrimin, P.; D’Angeli, F.; Veronese, A.C. Phase-Transfer-Catalyzed Reactions of a-Haloalmides: Synthesis of a-Lactams. Synthesis 1982, 1982, 586–587. [Google Scholar] [CrossRef]
- Lohman, D.; Edwards, D.; Wolfenden, R. Catalysis by desolvation: The catalytic prowess of SAM-dependent halide-alkylating enzymes. J. Am. Chem Soc. 2013, 135, 14473–14475. [Google Scholar] [CrossRef]
- Luo, Z.; Hou, J.; Menin, L.; Ong, Q.; Stellacci, F. Evolution of the Ligand Shell Morphology during Ligand Exchange Reactions on Gold Nanoparticles. Angew. Chem. Int. Ed. 2017, 56, 13521–13525. [Google Scholar] [CrossRef]
- Ong, Q.; Nianias, N.; Stellacci, F. A review of molecular phase separation in binary self-assembled monolayers of thiols on gold surfaces. Epl. Europhys. Lett. 2017, 119, 66001. [Google Scholar] [CrossRef]
- Ong, Q.; Luo, Z.; Stellacci, F. Characterization of Ligand Shell for Mixed-Ligand Coated Gold Nanoparticles. Accounts Chem Res. 2017, 50, 1911–1919. [Google Scholar] [CrossRef]
- Şologan, M.; Marson, D.; Polizzi, S.; Pengo, P.; Boccardo, S.; Pricl, S.; Posocco, P.; Pasquato, L. Patchy and Janus Nanoparticles by Self-Organization of Mixtures of Fluorinated and Hydrogenated Alkanethiolates on the Surface of a Gold Core. ACS Nano 2016, 10, 9316–9325. [Google Scholar] [CrossRef] [Green Version]
- Guarino, G.; Rastrelli, F.; Scrimin, P.; Mancin, F. Lanthanide-based NMR: A tool to investigate component distribution in mixed-monolayer-protected nanoparticles. J. Am. Chem. Soc. 2012, 134, 7200–7203. [Google Scholar] [CrossRef]
- Guarino, G.; Rastrelli, F.; Mancin, F. Mapping the nanoparticle-coating monolayer with NMR pseudocontact shifts. Chem. Commun. 2011, 48, 1523–1525. [Google Scholar] [CrossRef]
- Hoyle, C.E.; Bowman, C.N. Thiol–Ene Click Chemistry. Angew. Chem. Int. Ed. 2010, 49, 1540–1573. [Google Scholar] [CrossRef]
- Manea, F.; Bindoli, C.; Polizzi, S.; Lay, L.; Scrimin, P. Expeditious Synthesis of Water-Soluble, Monolayer-Protected Gold Nanoparticles of Controlled Size and Monolayer Composition. Langmuir 2008, 24, 4120–4124. [Google Scholar] [CrossRef]
- Zaupa, G.; Mora, C.; Bonomi, R.; Prins, L.J.; Scrimin, P. Catalytic self-assembled monolayers on Au nanoparticles: The source of catalysis of a transphosphorylation reaction. Chemistry. 2011, 17, 4879–4889. [Google Scholar] [CrossRef]
- Salvio, R.; Volpi, S.; Cacciapaglia, R.; Sansone, F.; Mandolini, L.; Casnati, A. Upper Rim Bifunctional cone-Calix[4]arenes Based on a Ligated Metal Ion and a Guanidinium Unit as DNAase and RNAase Mimics. J. Org. Chem. 2016, 81, 4728–4735. [Google Scholar] [CrossRef]
- Salvio, R.; Volpi, S.; Cacciapaglia, R.; Casnati, A.; Mandolini, L.; Sansone, F. Ribonuclease Activity of an Artificial Catalyst that Combines a Ligated Cu(II) Ion and a Guanidinium Group at the Upper Rim of a cone-Calix[4]arene Platform. J. Org. Chem. 2015, 80, 5887–5893. [Google Scholar] [CrossRef]
- Pezzato, C.; Scrimin, P.; Prins, L.J. Zn2+-regulated self-sorting and mixing of phosphates and carboxylates on the surface of functionalized gold nanoparticles. Angew. Chem. Int. Ed. Engl. 2014, 53, 2104–2109. [Google Scholar] [CrossRef]
- Bencze, E.; Zonta, C.; Mancin, F.; Prins, L.; Scrimin, P. Distance between Metal Centres Affects Catalytic Efficiency of Dinuclear CoIII Complexes in the Hydrolysis of a Phosphate Diester. Eur J. Org. Chem. 2018, 2018, 5375–5381. [Google Scholar] [CrossRef]
- Molenveld, P.; Kapsabelis, S.; Engbersen, J.; Reinhoudt, D. Highly Efficient Phosphate Diester Transesterification by a Calix[4 ]arene-Based Dinuclear Zinc(II) Catalyst. J. Am. Chem. Soc. 1997, 119, 2948–2949. [Google Scholar] [CrossRef]
- Scrimin, P.; Tecilla, P.; Tonellato, U. Leaving group effect in the cleavage of picolinate esters catalyzed by hydroxy-functionalized metallomicelles. J. Org. Chem. 1994, 59, 18–24. [Google Scholar] [CrossRef]
- Riccardi, L.; Gabrielli, L.; Sun, X.; De Biasi, F.; Rastrelli, F.; Mancin, F.; De Vivo, M. Nanoparticle-Based Receptors Mimic Protein-Ligand Recognition. Chem. 2017, 3, 92–109. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.; Anslyn, E. A cationic host displaying positive cooperativity in water. Proc. Natl. Acad. Sci. USA 2007, 104, 6538–6543. [Google Scholar] [CrossRef] [Green Version]
- Piatek, A.; Gray, M.; Anslyn, E. Guanidinium Groups Act as General-Acid Catalysts in Phosphoryl Transfer Reactions: A Two-Proton Inventory on a Model System. J. Am. Chem. Soc. 2004, 126, 9878–9879. [Google Scholar] [CrossRef]
- Feng, G.; Natale, D.; Prabaharan, R.; Mareque-Rivas, J.C.; Williams, N.H. Efficient phosphodiester binding and cleavage by a Zn(II) complex combining hydrogen-bonding interactions and double Lewis acid activation. Angew. Chem. Int. Ed. Engl. 2006, 45, 7056–7059. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds AuNP1-5 are available from the authors. |








| Catalyst | kcat, s−1, a | k2, l × mol−1 × s−1, b | KM, mM | krelc |
|---|---|---|---|---|
| AuNP1 | 0.193 | 331 | 0.58 | 9.7 × 105 |
| AuNP2 | 0.031 | 81.6 | 0.38 | 1.5 × 105 |
| AuNP3 | 0.010 | 9.4 | 1.1 | 5.3 × 104 |
| AuNP4 | 0.014 | 5.1 | 2.7 | 6.9 × 104 |
| AuNP5 | 0.006 | 3.85 | 1.7 | 3.3 × 104 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czescik, J.; Zamolo, S.; Darbre, T.; Mancin, F.; Scrimin, P. Factors Influencing the Activity of Nanozymes in the Cleavage of an RNA Model Substrate. Molecules 2019, 24, 2814. https://doi.org/10.3390/molecules24152814
Czescik J, Zamolo S, Darbre T, Mancin F, Scrimin P. Factors Influencing the Activity of Nanozymes in the Cleavage of an RNA Model Substrate. Molecules. 2019; 24(15):2814. https://doi.org/10.3390/molecules24152814
Chicago/Turabian StyleCzescik, Joanna, Susanna Zamolo, Tamis Darbre, Fabrizio Mancin, and Paolo Scrimin. 2019. "Factors Influencing the Activity of Nanozymes in the Cleavage of an RNA Model Substrate" Molecules 24, no. 15: 2814. https://doi.org/10.3390/molecules24152814