RNA-seq Based Transcriptome Analysis of the Anti-Obesity Effect of Green Tea Extract Using Zebrafish Obesity Models
Abstract
:1. Introduction
2. Results
2.1. GTE Decreased Visceral Adipose Tissue (VAT) Volume Induced by High-Fat Diet in Zebrafish Larvae
2.2. GTE Reduced VAT and Decreased Plasma TG and TCHO Levels in Adult Obese Zebrafish
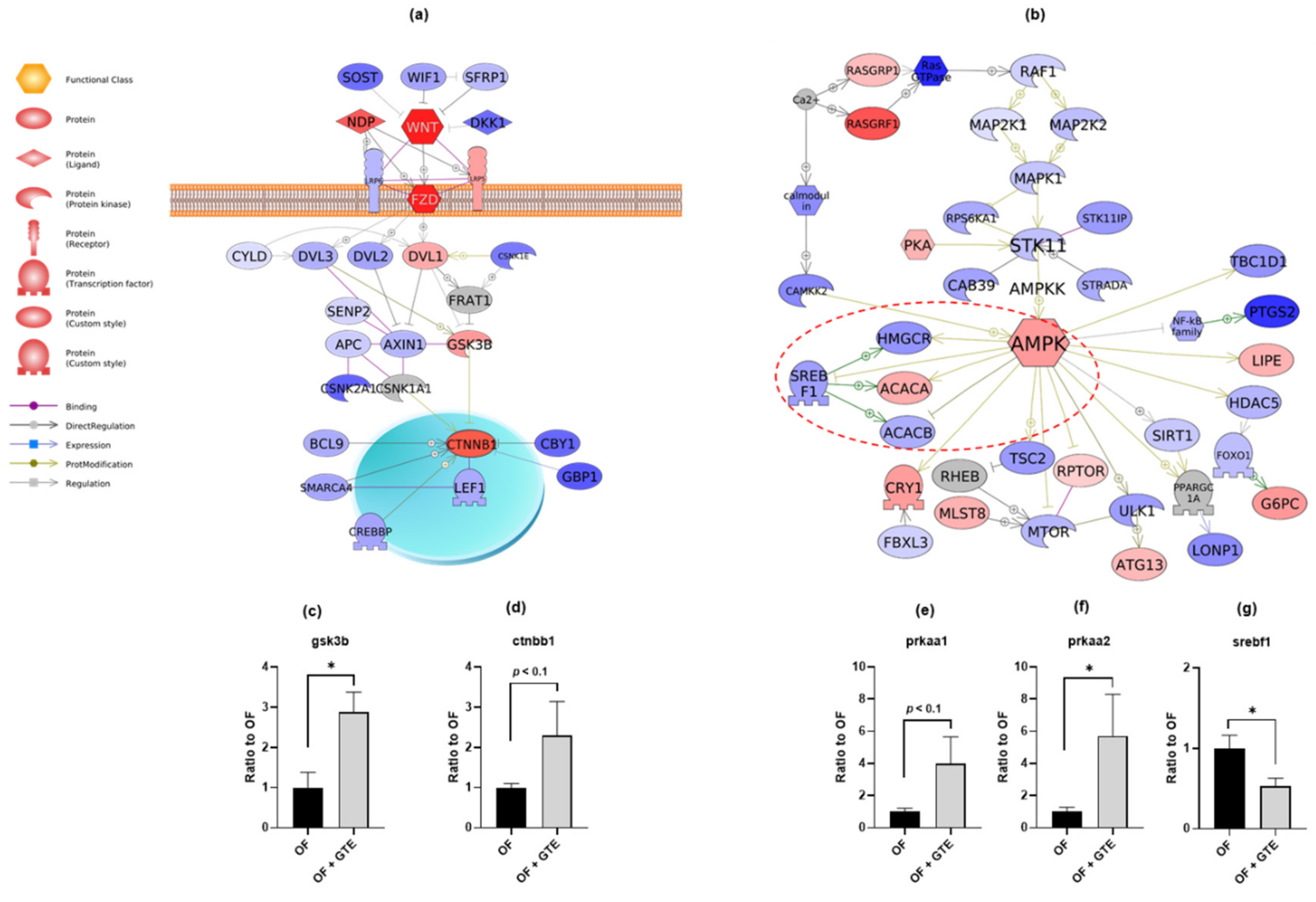
2.3. GTE Exhibits Anti-Obesity Effects via Wnt/β-catenin and AMPK Pathways
2.4. Common Pathways of the Anti-Obesity Mechanism in GTE between Zebrafish and Mouse
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Zebrafish Strains and Maintenance
4.3. Chemicals and Zebrafish Obesogenic Test in Zebrafish Larvae
4.4. Adult Zebrafish Treatment
4.5. CT Measurement of VAT Volume
4.6. RNA Isolation, Library Construction, and High-Throughput Sequencing
4.7. Bioinformatics Analysis of RNA-Seq data
4.8. Real-time Quantitative Polymerase Chain Reaction (qPCR) Analysis
4.9. Statistical Aalysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perva-Uzunalic, A.; Skerget, M.; Knez, Z.; Weinreich, B.; Otto, F.; Gruner, S. Extraction of active ingredients from green tea (Camellia sinensis): Extraction efficiency of major catechins and caffeine. Food Chem. 2006, 96, 597–605. [Google Scholar] [CrossRef]
- Sano, M.; Tabata, M.; Suzuki, M.; Degawa, M.; Miyase, T.; Maeda-Yamamoto, M. Simultaneous determination of twelve tea catechins by high-performance liquid chromatography with electrochemical detection. Analyst 2001, 126, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Buer, J.; Pietschmann, T.; Steinmann, E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013, 168, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Kaihatsu, K.; Yamabe, M.; Ebara, Y. Antiviral Mechanism of Action of Epigallocatechin-3-O-gallate and Its Fatty Acid Esters. Molecules 2018, 23, 2475. [Google Scholar] [CrossRef] [PubMed]
- Taguri, T.; Tanaka, T.; Kouno, I. Antibacterial spectrum of plant polyphenols and extracts depending upon hydroxyphenyl structure. Biol. Pharm. Bull. 2006, 29, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Bailey, H.H.; Mukhtar, H. Green tea polyphenols and cancer chemoprevention of genitourinary cancer. In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2013. [Google Scholar] [CrossRef]
- Saleem, M.; Adhami, V.M.; Siddiqui, I.A.; Mukhtar, H. Tea beverage in chemoprevention of prostate cancer: A mini-review. Nutr. Cancer 2003, 47, 13–23. [Google Scholar] [CrossRef]
- Matsumoto, N.; Ishigaki, F.; Ishigaki, A.; Iwashina, H.; Hara, Y. Reduction of Blood-Glucose Levels by Tea Catechin. Biosci. Biotechnol. Biochem 1993, 57, 525–527. [Google Scholar] [CrossRef]
- Sabu, M.C.; Smitha, K.; Ramadasan, K. Anti-diabetic activity of green tea polyphenols and their role in reducing oxidative stress in experimental diabetes. J. Ethnopharmacol. 2002, 83, 109–116. [Google Scholar]
- Legeay, S.; Rodier, M.; Fillon, L.; Faure, S.; Clere, N. Epigallocatechin Gallate: A Review of Its Beneficial Properties to Prevent Metabolic Syndrome. Nutrients 2015, 7, 5230. [Google Scholar] [CrossRef]
- Tijburg, L.B.M.; Mattern, T.; Folts, J.D.; Weisgerber, U.M.; Katan, M.B. Tea flavonoids and cardiovascular diseases: A review. Crit. Rev. Food Sci. 1997, 37, 771–785. [Google Scholar] [CrossRef]
- Suzuki, T.; Pervin, M.; Goto, S.; Isemura, M.; Nakamura, Y. Beneficial Effects of Tea and the Green Tea Catechin Epigallocatechin-3-gallate on Obesity. Molecules 2016, 21, 1305. [Google Scholar] [CrossRef] [PubMed]
- Isemura, M. Catechin in Human Health and Disease. Molecules 2019, 24, 528. [Google Scholar] [CrossRef] [PubMed]
- Hasumura, T.; Shimada, Y.; Kuroyanagi, J.; Nishimura, Y.; Meguro, S.; Takema, Y.; Tanaka, T. Green tea extract suppresses adiposity and affects the expression of lipid metabolism genes in diet-induced obese zebrafish. Nutr. Metab. 2012, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Nagao, T.; Hase, T.; Tokimitsu, I. A green tea extract high in catechins reduces body fat and cardiovascular risks in humans. Obesity 2007, 15, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, Y.; Xie, Z.; Zhou, Y.; Zhang, Y.; Wan, X. The anti-obesity effects of green tea in human intervention and basic molecular studies. Eur. J. Clin. Nutr. 2014, 68, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Freifeld, L.; Odstrcil, I.; Forster, D.; Ramirez, A.; Gagnon, J.A.; Randlett, O.; Costa, E.K.; Asano, S.; Celiker, O.T.; Gao, R.; et al. Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc. Natl. Acad. Sci. USA 2017, 114, E10799–E10808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dooley, K.; Zon, L.I. Zebrafish: A model system for the study of human disease. Curr. Opin. Genet. Dev. 2000, 10, 252–256. [Google Scholar] [CrossRef]
- McGrath, P.; Li, C.Q. Zebrafish: A predictive model for assessing drug-induced toxicity. Drug Discov. Today 2008, 13, 394–401. [Google Scholar] [CrossRef]
- Zang, L.; Maddison, L.A.; Chen, W. Zebrafish as a Model for Obesity and Diabetes. Front. Cell Dev. Biol. 2018, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Tingaud-Sequeira, A.; Ouadah, N.; Babin, P.J. Zebrafish obesogenic test: A tool for screening molecules that target adiposity. J. Lipid Res. 2011, 52, 1765–1772. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, Y.Q.; Guo, S.Y.; Li, C.Q. Rapid analysis of hypolipidemic drugs in a live zebrafish assay. J. Pharmacol. Toxicol. Methods 2015, 72, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.J., III; Trent, C.M.; Rawls, J.F. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J. Lipid Res. 2009, 50, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Nishimura, Y.; Zang, L.; Hirano, M.; Shimada, Y.; Wang, Z.; Umemoto, N.; Kuroyanagi, J.; Nishimura, N.; Tanaka, T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010, 10, 21. [Google Scholar] [CrossRef]
- Zang, L.; Shimada, Y.; Kawajiri, J.; Tanaka, T.; Nishimura, N. Effects of Yuzu (Citrus junos Siebold ex Tanaka) peel on the diet-induced obesity in a zebrafish model. J. Funct. Foods 2014, 10, 499–510. [Google Scholar] [CrossRef]
- Zang, L.Q.; Shimada, Y.; Tanaka, T.; Nishimura, N. Rhamnan sulphate from Monostroma nitidum attenuates hepatic steatosis by suppressing lipogenesis in a diet-induced obesity zebrafish model. J. Funct. Foods 2015, 17, 364–370. [Google Scholar] [CrossRef]
- Nakayama, H.; Shimada, Y.; Zang, L.; Terasawa, M.; Nishiura, K.; Matsuda, K.; Toombs, C.; Langdon, C.; Nishimura, N. Novel Anti-Obesity Properties of Palmaria mollis in Zebrafish and Mouse Models. Nutrients 2018, 10, 1401. [Google Scholar] [CrossRef] [PubMed]
- Kotelnikova, E.; Yuryev, A.; Mazo, I.; Daraselia, N. Computational approaches for drug repositioning and combination therapy design. J. Bioinform. Comput. Biol. 2010, 8, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Majewska, A.; Domoradzki, T.; Grzelkowska-Kowalczyk, K. Transcriptomic Profiling During Myogenesis. Methods Mol. Biol. 2019, 1889, 127–168. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Kuninaga, S.; Ariyoshi, M.; Zhang, B.; Shiina, Y.; Takahashi, Y.; Umemoto, N.; Nishimura, Y.; Enari, H.; Tanaka, T. E2F8 promotes hepatic steatosis through FABP3 expression in diet-induced obesity in zebrafish. Nutr. Metab. 2015, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Bae, S.; Yoon, Y. The anti-adipogenic effects of (−)epigallocatechin gallate are dependent on the WNT/beta-catenin pathway. J. Nutr. Biochem. 2013, 24, 1232–1240. [Google Scholar] [CrossRef]
- Li, F.; Gao, C.; Yan, P.; Zhang, M.; Wang, Y.; Hu, Y.; Wu, X.; Wang, X.; Sheng, J. EGCG Reduces Obesity and White Adipose Tissue Gain Partly Through AMPK Activation in Mice. Front. Pharmacol. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, A.; Nandy, S.K.; Sarkar, J.; Chakraborti, T.; Chakraborti, S. Inhibition of pro-/active MMP-2 by green tea catechins and prediction of their interaction by molecular docking studies. Mol. Cell. Biochem. 2017, 427, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Benyon, R.C.; Iredale, J.P.; Goddard, S.; Winwood, P.J.; Arthur, M.J. Expression of tissue inhibitor of metalloproteinases 1 and 2 is increased in fibrotic human liver. Gastroenterology 1996, 110, 821–831. [Google Scholar] [CrossRef]
- Grigorescu, M. Noninvasive biochemical markers of liver fibrosis. J. Gastrointest. Liver Dis. JGLD 2006, 15, 149–159. [Google Scholar]
- Lefebvre, V.; Dvir-Ginzberg, M. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect. Tissue Res. 2017, 58, 2–14. [Google Scholar] [CrossRef]
- Stockl, S.; Bauer, R.J.; Bosserhoff, A.K.; Gottl, C.; Grifka, J.; Grassel, S. Sox9 modulates cell survival and adipogenic differentiation of multipotent adult rat mesenchymal stem cells. J. Cell Sci. 2013, 126, 2890–2902. [Google Scholar] [CrossRef] [Green Version]
- Hudak, C.S.; Sul, H.S. Pref-1, a gatekeeper of adipogenesis. Front. Endocrinol. 2013, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, Q.; Liu, L.; Wu, H.; Zheng, L. Effect of epigallocatechin-3-gallate on proliferation and phenotype maintenance in rabbit articular chondrocytes in vitro. Exp. Ther. Med. 2015, 9, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book, 5th ed.; A Guide for the Laboratory Use of Zebrafish (Danio rerio); University of Oregon Press: Eugene, OR, USA, 2007. [Google Scholar]
- Zang, L.; Morikane, D.; Shimada, Y.; Tanaka, T.; Nishimura, N. A novel protocol for the oral administration of test chemicals to adult zebrafish. Zebrafish 2011, 8, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.Q.; Shimada, Y.; Nishimura, Y.; Tanaka, T.; Nishimura, N. A Novel, Reliable Method for Repeated Blood Collection from Aquarium Fish. Zebrafish 2013, 10, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.Q.; Shimada, Y.; Nishimura, Y.; Tanaka, T.; Nishimura, N. Repeated Blood Collection for Blood Tests in Adult Zebrafish. J. Vis. Exp. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.; Shimada, Y.; Nishimura, N. Development of a Novel Zebrafish Model for Type 2 Diabetes Mellitus. Sci. Rep. 2017, 7, 1461. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Green tea extract is available from the authors. |



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zang, L.; Shimada, Y.; Nakayama, H.; Kim, Y.; Chu, D.-C.; Juneja, L.R.; Kuroyanagi, J.; Nishimura, N. RNA-seq Based Transcriptome Analysis of the Anti-Obesity Effect of Green Tea Extract Using Zebrafish Obesity Models. Molecules 2019, 24, 3256. https://doi.org/10.3390/molecules24183256
Zang L, Shimada Y, Nakayama H, Kim Y, Chu D-C, Juneja LR, Kuroyanagi J, Nishimura N. RNA-seq Based Transcriptome Analysis of the Anti-Obesity Effect of Green Tea Extract Using Zebrafish Obesity Models. Molecules. 2019; 24(18):3256. https://doi.org/10.3390/molecules24183256
Chicago/Turabian StyleZang, Liqing, Yasuhito Shimada, Hiroko Nakayama, Youngil Kim, Djong-Chi Chu, Lekh Raj Juneja, Junya Kuroyanagi, and Norihiro Nishimura. 2019. "RNA-seq Based Transcriptome Analysis of the Anti-Obesity Effect of Green Tea Extract Using Zebrafish Obesity Models" Molecules 24, no. 18: 3256. https://doi.org/10.3390/molecules24183256




