Accramycin A, A New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
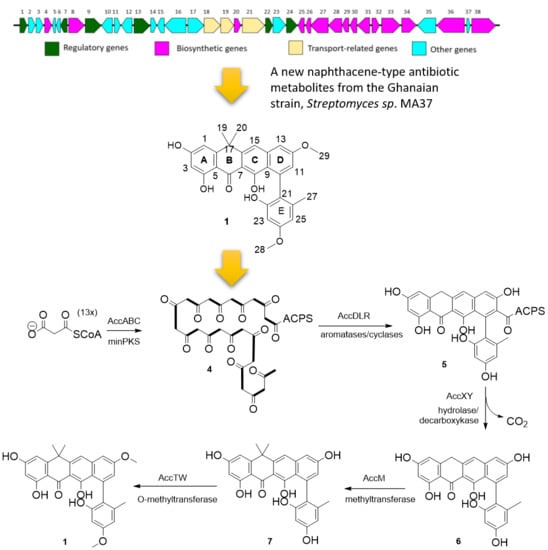
2.2. Proposed acc Biosynthetic Gene Cluster and Pathway
2.3. Antibacterial Assay
3. Materials and Methods
3.1. Fermentation
3.2. Extraction and Isolation
3.3. MS/MS Molecular Networking
3.4. Spectroscopic Analysis
3.5. Antibacterial Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [PubMed]
- Welsch, M.E.; Snyder, S.A.; Stockwell, B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010, 14, 347–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Kallifidas, D.; Brady, S.F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl. Acad. Sci. 2011, 108, 12629–12634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Chakraborty, D.; Dewell, S.B.; Reddy, B.V.B.; Brady, S.F. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J. Am. Chem. Soc. 2012, 134, 2981–2987. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, A.; Kim, Y.P.; Iwatsuki, M.; Hirose, T.; Sunazuka, T.; Hanaki, H.; Omura, S.; Shiomi, K. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. II. Structure elucidation. J. Antibiot. (Tokyo) 2017, 70, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, A.; Kim, Y.P.; Matsumoto, A.; Takahashi, Y.; Suzuki, M.; Onodera, H.; Tomoda, H.; Matsui, H.; Hanaki, H.; Iwatsuki, M.; et al. Naphthacemycins, novel circumventors of β-lactam resistance in MRSA, produced by Streptomyces sp. KB-3346-5. I. The taxonomy of the producing strain, and the fermentation, isolation and antibacterial activities. J. Antibiot. (Tokyo) 2017, 70, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Kojima, Y.; Matsui, H.; Hanaki, H.; Iwatsuki, M.; Shiomi, K.; Omura, S.; Sunazuka, T. Total synthesis of (±)-naphthacemycin A 9, possessing both antibacterial activity against methicillin-resistant Staphylococcus aureus and circumventing effect of β-lactam resistance. J. Antibiot. (Tokyo) 2017, 70, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tabudravu, J.; Elsayed, S.S.; Travert, J.; Peace, D.; Tong, M.H.; Kyeremeh, K.; Kelly, S.M.; Trembleau, L.; Ebel, R.; et al. Discovery of a single monooxygenase that catalyzes carbamate formation and ring contraction in the biosynthesis of the legonmycins. Angew. Chemie - Int. Ed. 2015, 54, 12697–12701. [Google Scholar] [CrossRef]
- Su, L.; Lv, M.; Kyeremeh, K.; Deng, Z.; Deng, H.; Yu, Y. A ThDP-dependent enzymatic carboligation reaction involved in Neocarazostatin A tricyclic carbazole formation. Org. Biomol. Chem. 2016, 14, 8679–8684. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Zhang, R.; Kyeremeh, K.; Deng, Z.; Deng, H.; Yu, Y. Dissection of the neocarazostatin: a C 4 alkyl side chain biosynthesis by in vitro reconstitution. Org. Biomol. Chem. 2017, 3843–3848. [Google Scholar] [CrossRef]
- Huang, S.; Elsayed, S.S.; Lv, M.; Tabudravu, J.; Rateb, M.E.; Gyampoh, R.; Kyeremeh, K.; Ebel, R.; Jaspars, M.; Deng, Z.; et al. Biosynthesis of Neocarazostatin A Reveals the Sequential Carbazole Prenylation and Hydroxylation in the Tailoring Steps. Chem. Biol. 2015, 22, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Maglangit, F.; Tong, M.H.; Jaspars, M.; Kyeremeh, K.; Deng, H. Legonoxamines A-B, two new hydroxamate siderophores from the soil bacterium, Streptomyces sp. MA37. Tetrahedron Lett. 2019, 60, 75–79. [Google Scholar] [CrossRef]
- Deng, H.; Ma, L.; Bandaranayaka, N.; Qin, Z.; Mann, G.; Kyeremeh, K.; Yu, Y.; Shepherd, T.; Naismith, J.H.; O’Hagan, D. Identification of Fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by Genome Mining. ChemBioChem 2014, 15, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bartholome, A.; Tong, M.H.; Qin, Z.; Yu, Y.; Shepherd, T.; Kyeremeh, K.; Deng, H.; O’Hagan, D.; Su, L.; et al. Identification of a fluorometabolite from Streptomyces sp. MA37: (2R3S4S)-5-fluoro-2,3,4-trihydroxypentanoic acid. Chem. Sci. 2017, 14, 8679–8684. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: different manners, one goal. Nat. Prod. Rep. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tabudravu, J.N.; Pellissier, L.; Smith, A.J.; Subko, K.; Autréau, C.; Feussner, K.; Hardy, D.; Butler, D.; Kidd, R.; Milton, E.J.; et al. LC-HRMS-Database screening metrics for rapid prioritization of samples to accelerate the discovery of structurally new natural products. J. Nat. Prod. 2019, 82, 211–220. [Google Scholar] [CrossRef]
- Chervin, J.; Stierhof, M.; Tong, M.H.; Peace, D.; Hansen, K.Ø.; Urgast, D.S.; Andersen, J.H.; Yu, Y.; Ebel, R.; Kyeremeh, K.; et al. Targeted Dereplication of Microbial Natural Products by High-Resolution MS and Predicted LC Retention Time. J. Nat. Prod. 2017, 80, 1370–1377. [Google Scholar] [CrossRef]
- Qin, Z.; Munnoch, J.T.; Devine, R.; Holmes, N.A.; Seipke, R.F.; Wilkinson, K.A.; Wilkinson, B.; Hutchings, M.I. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem. Sci. 2017, 8, 3218–3227. [Google Scholar] [CrossRef] [Green Version]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Kudo, F.; Yonezawa, T.; Komatsubara, A.; Mizoue, K.; Eguchi, T. Cloning of the biosynthetic gene cluster for naphthoxanthene antibiotic FD-594 from Streptomyces sp. TA-0256. J. Antibiot. (Tokyo) 2011, 64, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Chen, H.; Guo, Z.; Liu, N.; Li, J.; Huang, Y.; Xiang, W.; Chen, Y. Discovery of pentangular polyphenols hexaricins A–C from marine Streptosporangium sp. CGMCC 4.7309 by genome mining. Appl. Microbiol. Biotechnol. 2016, 100, 4189–4199. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Naganawa, H.; Muraoka, Y.; Aoyagi, T.; Takeuchi, T. Biosynthesis of benastatin A. J. Antibiot. (Tokyo) 1992, 45, 1767–1772. [Google Scholar] [CrossRef] [PubMed]
- Jakobi, K.; Hertweck, C. A Gene Cluster Encoding Resistomycin Biosynthesis in Streptomyces resistomycificus; Exploring Polyketide Cyclization beyond Linear and Angucyclic Patterns. J. Am. Chem. Soc. 2004, 126, 2298–2299. [Google Scholar] [CrossRef]
- Wyche, T.P.; Piotrowski, J.S.; Hou, Y.; Braun, D.; Deshpande, R.; McIlwain, S.; Ong, I.M.; Myers, C.L.; Guzei, I.A.; Westler, W.M.; et al. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J. Antibiot. (Tokyo) 2017, 134, 1415–1427. [Google Scholar]
- Lam, Y.K.T.; Hensens, O.; Helms, G.; Williams, D.; Nallin, M.; Smith, J.; Gartner, S.; Rodrigueztt, L.H.; Stevens-miles, S. L-755,805, A New Polyketide Endothelin Binding Inhibitor from an Actinomycete. Tetrahedron Lett. 1995, 36, 2013–2016. [Google Scholar] [CrossRef]
- Xu, Z.; Schenk, A.; Hertweck, C. Molecular analysis of the benastatin biosynthetic pathway and genetic engineering of altered fatty acid-polyketide hybrids. J. Am. Chem. Soc. 2007, 129, 6022–6030. [Google Scholar] [CrossRef]
- Nanduri, S.A.; Petit, S.; Smelser, C.; Apostol, M.; Alden, N.B.; Harrison, L.H.; Lynfield, R.; Vagnone, P.S.; Burzlaff, K.; Spina, N.L.; et al. Epidemiology of Invasive Early-Onset and Late-Onset Group B Streptococcal Disease in the United States, 2006 to 2015. JAMA Pediatr. 2008, 299, 2056–2065. [Google Scholar] [CrossRef] [PubMed]
- Skoff, T.H.; Farley, M.M.; Petit, S.; Craig, A.S.; Schaffner, W.; Gershman, K.; Harrison, L.H.; Lynfield, R.; Mohle-Boetani, J.; Zansky, S.; et al. Increasing Burden of Invasive Group B Streptococcal Disease in Nonpregnant Adults, 1990–2007. Clin. Infect. Dis. 2009, 49, 85–92. [Google Scholar] [CrossRef]
- Schrag, S.J.; Zywicki, S.; Farley, M.M.; Reingold, A.L.; Harrison, L.H.; Harrison, L.H.; Lefkowitz, L.; Hadler, J.L.; Danila, R.; Cieslak, P.; et al. Group B Streptococcal Disease in the WEra of Intrapartum Antibiotic Prophylaxis. N. Engl. J. Med. 2000, 342, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Blancas, D.; Santin, M.; Olmo, M.; Alcaide, F.; Carratala, J.; Gudiol, F. Group B streptococcal disease in nonpregnant adults: Incidence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Farley, M.M. Group B Streptococcal Disease in Nonpregnant Adults. Clin. Infect. Dis. 2002, 33, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Berg, B.R.; Houseman, J.L.; TerSteeg, Z.E.; LeBar, W.D.; Newton, D.W. Antimicrobial susceptibilities of group B streptococcus isolates from prenatal screening samples. J. Clin. Microbiol. 2014, 52, 3499–3501. [Google Scholar] [CrossRef] [PubMed]
- Lamagni, T.L.; Keshishian, C.; Efstratiou, A.; Guy, R.; Henderson, K.L.; Broughton, K.; Sheridan, E. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clin. Infect. Dis. 2013, 57, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Heelan, J.S.; Hasenbein, M.E.; McAdam, A.J. Resistance of Group B Streptococcus to Selected Antibiotics, Including Erythromycin and Clindamycin. J. Clin. Microbiol. 2004, 42, 1263–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.; Nichols, M.; Schrag, S.J. Two Cases of Invasive Vancomycin-Resistant Group B Streptococcus Infection. N. Engl. J. Med. 2014, 370, 885–886. [Google Scholar] [CrossRef] [PubMed]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr Protoc Bioinforma. 2014, 46, 1–7. [Google Scholar]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; De Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing Natural Product Diversity in a Collection of 146 Bacterial Strains Based on Growth and Extraction Protocols. J. Nat. Prod. 2017, 80, 588–597. [Google Scholar] [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007, 2, 2366–2382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A.; et al. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016, 28, 1–12. [Google Scholar] [CrossRef]
Sample Availability: Not available. |


| Position | 13C a | 1H, mult. (J, Hz) | Position | 13C a | 1H, mult. (J, Hz) |
|---|---|---|---|---|---|
| 1 | 112.1, CH | 6.49, d (2.4) | 16 | 147.4, C | - |
| 2 | 167.0, C b | - | 17 | 39.3, C | - |
| 3 | 104.0, CH | 5.99, d (2.4) | 18 | 154.5, C | - |
| 4 | 167.1, C | - | 19 | 34.4, CH3 | 1.70, s |
| 5 | 105.0, C | - | 20 | 34.7, CH3 | 1.69, s |
| 6 | 189.2, C | - | 21 | 125.3, C | - |
| 7 | 107.4, C | - | 22 | 155.2, C | - |
| 8 | 165.3, C | - | 23 | 99.2, CH | 6.32, d (2.4) |
| 9 | 118.7, C | - | 24 | 160.2, C | - |
| 10 | 141.0, C b | - | 25 | 106.9, CH | 6.37, d (2.4) |
| 11 | 121.7, CH | 6.71, d (2.4) | 26 | 138.2, C | - |
| 12 | 161.1, C | - | 27 | 20.6, CH3 | 1.91, s |
| 13 | 106.6, CH | 7.19, d (2.4) | 28 | 55.3, CH3 | 3.80, s |
| 14 | 141.8, C | - | 29 | 55.6, CH3 | 3.95, s |
| 15 | 115.8, CH | 7.47, s |
| No. | Gene | AA | Deduced Function | No. | Gene | AA | Deduced Function |
|---|---|---|---|---|---|---|---|
| 1 | accE | 242 | DNA binding response regulator | 20 | accL | 119 | Type II PKS cyclase |
| 2 | orf1 | 226 | Synthase | 21 | accK | 509 | Na+/H+ exchanger |
| 3 | orf2 | 210 | Glycosyl transferase | 22 | accJ | 145 | MarR family transcriptional regulator |
| 4 | accM | 220 | SAM-dependent methyl transferase | 23 | accG | 359 | Sensor histidine kinase |
| 5 | orf3 | 60 | Hypothetical protein | 24 | accF | 227 | LuxR family response regulator |
| 6 | orf4 | 38 | Unknown | 25 | accD | 152 | Polyketide cyclase |
| 7 | accN | 112 | Transcriptional regulator | 26 | accC | 97 | Acyl carrier protein |
| 8 | accO | 305 | NAD(P)-dependent oxidoreductase | 27 | accB | 415 | Beta-ketoacyl synthase |
| 9 | accP | 275 | MerR family transcriptional regulator | 28 | accA | 426 | Beta-ketoacyl synthase |
| 10 | orf5 | 193 | Nucleoside phosphorylase | 29 | accR | 131 | Cupin (cyclase/monooxygenase) |
| 11 | orf6 | 146 | VOC family protein | 30 | accS | 113 | Antibiotic biosynthesis monooxygenase |
| 12 | orf7 | 202 | Isomerase | 31 | accT | 350 | O-methyl transferase |
| 13 | accI | 303 | LysR family transcriptional regulator | 32 | accU | 113 | Antibiotic biosynthesis monooxygenase |
| 14 | orf8 | 108 | Hypothetical protein | 33 | accV | 430 | Halogenase |
| 15 | orf9 | 140 | Hypothetical protein | 34 | accW | 348 | O-methyl transferase |
| 16 | orf10 | 386 | Molybdopterin-binding protein | 35 | orf12 | 480 | Phenylalanine specific permease |
| 17 | orf11 | 480 | domain containing proteins | 36 | accX | 777 | hydrolase |
| 18 | accH | 491 | ABC transporter | 37 | orf13 | 43 | DUF-1232 domain containing protein |
| 19 | accI | 319 | ABC transporter | 38 | accY | 517 | decarboxylase |
| Pathogen | MIC (µg/mL) |
|---|---|
| Streptococcus B. ATCC 12386 | 27 |
| Escherichia coli ATCC 25922 | >50 |
| Pseudomonas aeruginosa ATCC 27853 | >50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maglangit, F.; Fang, Q.; Leman, V.; Soldatou, S.; Ebel, R.; Kyeremeh, K.; Deng, H. Accramycin A, A New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37. Molecules 2019, 24, 3384. https://doi.org/10.3390/molecules24183384
Maglangit F, Fang Q, Leman V, Soldatou S, Ebel R, Kyeremeh K, Deng H. Accramycin A, A New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37. Molecules. 2019; 24(18):3384. https://doi.org/10.3390/molecules24183384
Chicago/Turabian StyleMaglangit, Fleurdeliz, Qing Fang, Valentin Leman, Sylvia Soldatou, Rainer Ebel, Kwaku Kyeremeh, and Hai Deng. 2019. "Accramycin A, A New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37" Molecules 24, no. 18: 3384. https://doi.org/10.3390/molecules24183384
APA StyleMaglangit, F., Fang, Q., Leman, V., Soldatou, S., Ebel, R., Kyeremeh, K., & Deng, H. (2019). Accramycin A, A New Aromatic Polyketide, from the Soil Bacterium, Streptomyces sp. MA37. Molecules, 24(18), 3384. https://doi.org/10.3390/molecules24183384