Identification of The Fipronil Resistance Associated Mutations in Nilaparvata lugens GABA Receptors by Molecular Modeling
Abstract
:1. Introduction
2. Results and Discussion
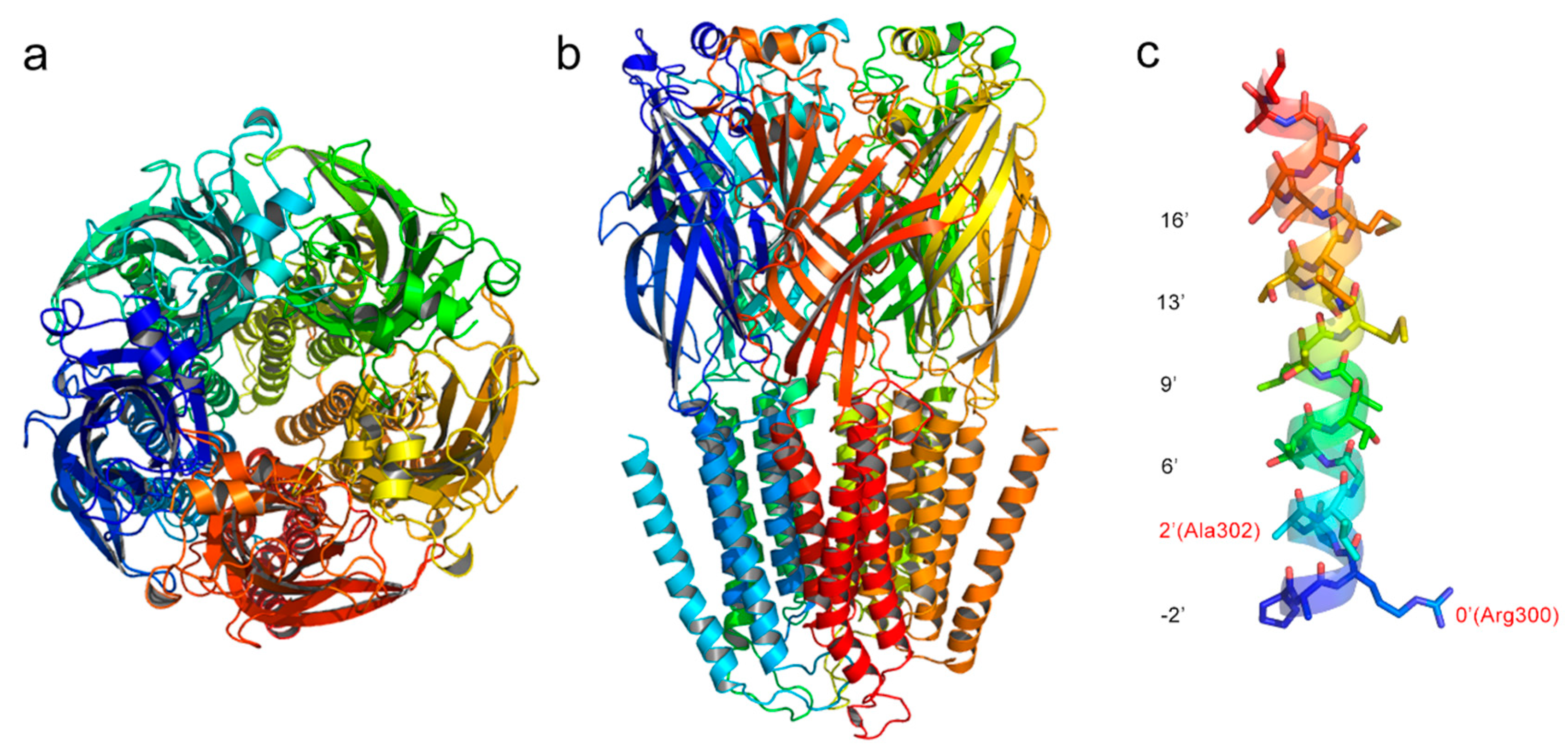
2.1. Homology Model
2.2. Model Optimization
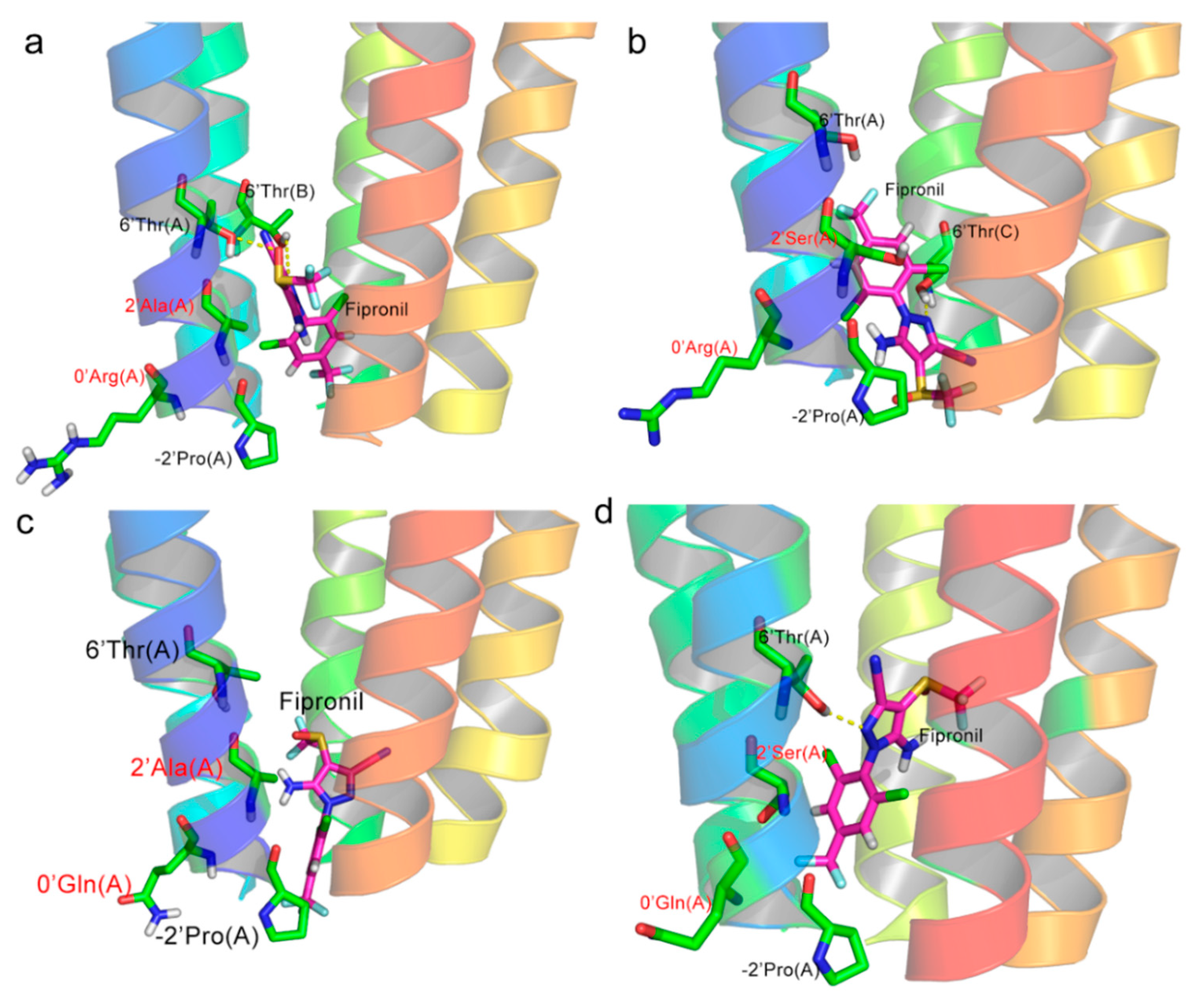
2.3. Ligand Docking
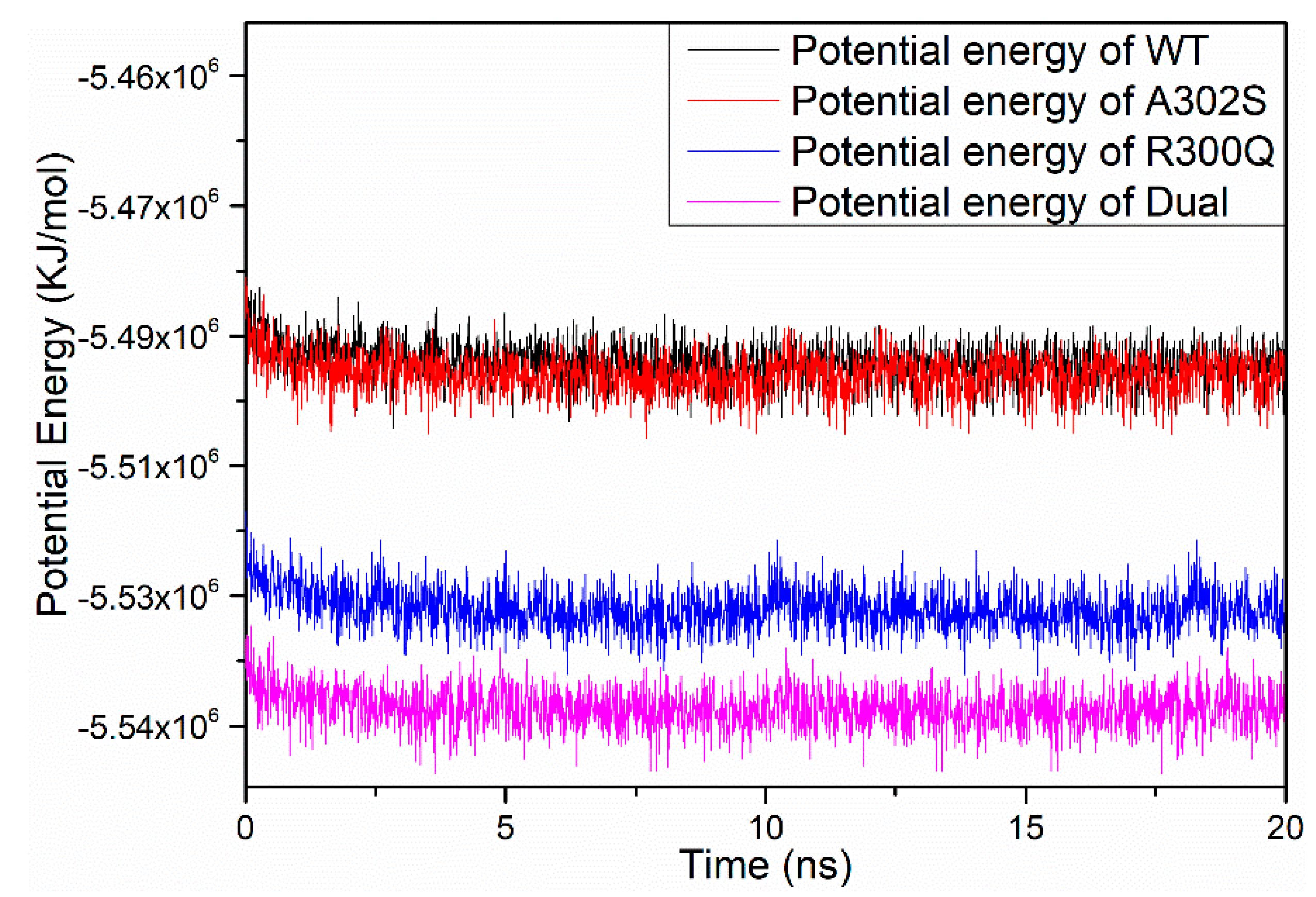
2.4. Molecular Dynamics Simulations
3. Materials and Methods
3.1. Protein Homology Modeling and Evaluation
3.2. Model Optimization
3.3. Docking Analysis
3.4. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GABA | γ-aminobutyric acid |
| GABAR | γ-aminobutyric acid receptor |
| CNS | Central nervous system |
| NCA | Noncompetitive antagonist |
| RDL | Resistant to dieldrin |
| WT | Wild type |
| MD | Molecular dynamics |
| 3D | Three-dimension |
| PE | Potential energy |
| RDLR | RDL GABA receptors |
| Rg | Radius of gyration |
| RMSD | Root-mean-square deviation |
| RMSF | Root mean square fluctuation |
| MSA | Multiple sequence alignment |
References
- Jager, K.W. Pesticides and Health. (Book Reviews: Aldrin, Dieldrin, Endrin and Telodrin. An Epidemiological and Toxicological Study of Long-Term Occupational Exposure). Science 1972, 172, 1323–1326. [Google Scholar]
- Ranson, H.; Rossiter, L.; Ortelli, F.; Jensen, B.; Wang, X.; Roth, C.W.; Collins, F.H.; Hemingway, J. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 2001, 359, 295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, L.; He, L.; Yang, X.; Shi, Y.; Liao, S.; Yang, S.; Cheng, J.; Ren, T. Interactions of Fipronil within Fish and Insects: Experimental and Molecular Modeling Studies. J. Agric. Food Chem. 2018, 66, 5756–5761. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.S.; Smart, T.G. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010, 31, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, L.D.; Pecci, A.; Estrin, D.A. In Search of GABAA Receptor’s Neurosteroid Binding Sites. J. Med. Chem. 2019, 62, 5250–5260. [Google Scholar] [CrossRef]
- Charnet, P.; Labarca, C.; Leonard, R.J.; Vogelaar, N.J.; Czyzyk, L.; Gouin, A.; Davidson, N.; Lester, H.A. An open-channel blocker interacts with adjacent turns of α-helices in the nicotinic acetylcholine receptor. Neuron 1990, 4, 87–95. [Google Scholar] [CrossRef]
- Perret, P.; Sarda, X.; Wolff, M.; Wu, T.; Bushey, D.; Goeldner, M. Interaction of non-competitive blockers within the γ-aminobutyric acid type A chloride channel using chemically reactive probes as chemical sensors for cysteine mutants. J. Biol. Chem. 1999, 274, 25350–25354. [Google Scholar] [CrossRef]
- Kliot, A.; Ghanim, M. Fitness costs associated with insecticide resistance. Pest. Manage. Sci. 2012, 68, 1431–1437. [Google Scholar] [CrossRef]
- Hope, M.; Menzies, M.; Kemp, D. Identification of a dieldrin resistance-associated mutation in Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). J. Econ. Entomol. 2010, 103, 1355–1359. [Google Scholar] [CrossRef]
- Martinez-Torres, D.; Foster, S.; Field, L.; Devonshire, A.; Williamson, M. A sodium channel point mutation is associated with resistance to DDT and pyrethroid insecticides in the peach-potato aphid, Myzus persicae (Sulzer)(Hemiptera: Aphididae). Insect Mol. Biol. 1999, 8, 339–346. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.; Mortlock, D.; Shaffer, C.; MacIntyre, R.; Roush, R.T. Molecular cloning and transformation of cyclodiene resistance in Drosophila: An invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc. Natl. Acad. Sci. USA 1991, 88, 7209–7213. [Google Scholar] [CrossRef] [PubMed]
- Domingues, L.N.; Guerrero, F.D.; Becker, M.E.; Alison, M.W.; Foil, L.D. Discovery of the Rdl mutation in association with a cyclodiene resistant population of horn flies, Haematobia irritans (Diptera: Muscidae). Vet. Parasitol. 2013, 198, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Awolola, T.; Howell, P.; Koekemoer, L.; Brooke, B.; Benedict, M.; Coetzee, M.; Zheng, L. Independent mutations in the Rdl locus confer dieldrin resistance to Anopheles gambiae and An. arabiensis. Insect Mol. Biol. 2005, 14, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Rocheleau, T.A.; Steichen, J.C.; Chalmers, A.E. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature 1993, 363, 449. [Google Scholar]
- Buckingham, S.D.; Biggin, P.C.; Sattelle, B.M.; Brown, L.A.; Sattelle, D.B. Insect GABA receptors: Splicing, editing, and targeting by antiparasitics and insecticides. Mol. Pharmacol. 2005, 68, 942–951. [Google Scholar] [CrossRef]
- Zhang, H.G.; ffrench-Constant, R.H.; Jackson, M.B. A unique amino acid of the Drosophila GABA receptor with influence on drug sensitivity by two mechanisms. J. Physiol. 1994, 479, 65–75. [Google Scholar] [CrossRef]
- Hosie, A.M.; Baylis, H.A.; Buckingham, S.D.; Sattelle, D.B. Actions of the insecticide fipronil, on dieldrin-sensitive and-resistant GABA receptors of Drosophila melanogaster. Br. J. Pharmacol. 1995, 115, 909–912. [Google Scholar] [CrossRef]
- Buckingham, S.; Matsuda, K.; Hosie, A.; Baylis, H.; Squire, M.; Lansdell, S.; Millar, N.; Sattelle, D. Wild-type and insecticide-resistant homo-oligomeric GABA receptors of Drosophila melanogaster stably expressed in a Drosophila cell line. Neuropharmacology 1996, 35, 1393–1401. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, X.; Yang, Y.; Li, H.; Wang, X.; Yang, B.; Zhang, J.; Li, C.; Millar, N.S.; Liu, Z. Synergistic and compensatory effects of two point mutations conferring target-site resistance to fipronil in the insect GABA receptor RDL. Sci. Rep. 2016, 6, 32335. [Google Scholar] [CrossRef]
- Wang, W.; Tian, Y.; Wan, Y.; Gu, S.; Ju, X.; Luo, X.; Liu, G. Insights into the key structural features of N 1-ary-benzimidazols as HIV-1 NNRTIs using molecular docking, molecular dynamics, 3D-QSAR, and pharmacophore modeling. Struct. Chem. 2019, 30, 385–397. [Google Scholar] [CrossRef]
- Wan, Y.; Tian, Y.; Wang, W.; Gu, S.; Ju, X.; Liu, G. In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using docking-based 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches. RSC Adv. 2018, 8, 40529–40543. [Google Scholar] [CrossRef]
- Liu, G.; Li, H.; Shi, J.; Wang, W.; Furuta, K.; Liu, D.; Zhao, C.; Ozoe, F.; Ju, X.; Ozoe, Y. 4-Aryl-5-carbamoyl-3-isoxazolols as competitive antagonists of insect GABA receptors: Synthesis, biological activity, and molecular docking studies. Bioorg. Med. Chem. 2019, 27, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, W.; Wan, Y.; Ju, X.; Gu, S. Application of 3D-QSAR, pharmacophore, and molecular docking in the molecular design of diarylpyrimidine derivatives as HIV-1 nonnucleoside reverse transcriptase inhibitors. Int. J. Mol. Sci. 2018, 19, 1436. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wan, Y.; Wang, W.; Fang, S.; Gu, S.; Ju, X. Docking-based 3D-QSAR and pharmacophore studies on diarylpyrimidines as non-nucleoside inhibitors of HIV-1 reverse transcriptase. Mol. Divers. 2019, 23, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Zheng, N.; Cheng, J.; Zhang, W.; Li, W.; Shao, X.; Xu, Z.; Xu, X.; Li, Z. Binding difference of fipronil with GABAARs in fruitfly and zebrafish: Insights from homology modeling, docking, and molecular dynamics simulation studies. J. Agric. Food Chem. 2014, 62, 10646–10653. [Google Scholar] [CrossRef]
- Cheng, J.; Ju, X.; Chen, X.; Liu, G. Homology modeling of human α1β2γ2 and house fly β3 GABA receptor channels and Surflex-docking of fipronil. J. Mol. Model. 2009, 15, 1145–1153. [Google Scholar] [CrossRef]
- Ci, S.; Ren, T.; Su, Z. Modeling the interaction of fipronil-related non-competitive antagonists with the GABA β3-receptor. J. Mol. Model. 2007, 13, 457. [Google Scholar] [CrossRef]
- Chen, L.; Durkin, K.A.; Casida, J.E. Structural model for γ-aminobutyric acid receptor noncompetitive antagonist binding: Widely diverse structures fit the same site. Proc. Natl. Acad. Sci. USA 2006, 103, 5185–5190. [Google Scholar] [CrossRef]
- Nakao, T.; Naoi, A.; Kawahara, N.; Hirase, K. Mutation of the GABA receptor associated with fipronil resistance in the whitebacked planthopper, Sogatella furcifera. Pestic. Biochem. Physiol. 2010, 97, 262–266. [Google Scholar] [CrossRef]
- Casida, J.E.; Tomizawa, M. Insecticide interactions with γ-aminobutyric acid and nicotinic receptors: Predictive aspects of structural models. J. Pestic. Sci. 2008, 33, 4–8. [Google Scholar] [CrossRef]
- Hisano, K.; Ozoe, F.; Huang, J.; Kong, X.; Ozoe, Y. The channel-lining 6′ amino acid in the second membrane-spanning region of ionotropic GABA receptors has more profound effects on 4′-ethynyl-4-n-propylbicycloorthobenzoate binding than the 2′ amino acid. Invertebr. Neurosci. 2007, 7, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Handoko, S.D.; Kwoh, C.K. Cscore: A simple yet effective scoring function for protein–ligand binding affinity prediction using modified Cmac learning architecture. J. Bioinf. Comput. Biol. 2011, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABA A receptor. Nature 2014, 512, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, Y.; Tian, Y.; Zhao, Y.; Wu, F.; Luo, X.; Liu, G. In silico study of 3-hydroxypyrimidine-2, 4-diones as inhibitors of HIV RT-associated RNase H using molecular docking, molecular dynamics, 3D-QSAR, and pharmacophore model. New J. Chem. 2019, 43, 17004–17017. [Google Scholar] [CrossRef]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Da Silva, A.W.S.; Vranken, W.F. ACPYPE-Antechamber python parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
Sample Availability: Data are available from the corresponding author upon reasonable request. |



| Methods | Ramachandran Plot a | Z-Score b | Percentage of the Residues have Averaged 3D-1D Score ≥ 0.2 c | ERRAT (Overall Quality Factor) d | |
|---|---|---|---|---|---|
| Allowed Regions | Disallowed Regions | ||||
| SWISS-MODEL | 99.5% | 0.5% | −4.85 | 70.61% | 89.76 |
| MODELLER | 99.8% | 0.2% | −4.74 | 70.25% | 75.91 |
| SYBYL | 99.3% | 0.7% | −3.16 | 73.35% | 79.00 |
| Model | Hydrogen Bonds | Total Score | ΔGbinding (KJ/mol) | IC50 a |
|---|---|---|---|---|
| WT | 6′Thr(A) with Fipronil | 3.311 | −18.901 | 19.81 ± 3.31 |
| A302S | 6′Thr(C) with Fipronil | 3.59 | −20.494 | 45.47 ± 7.05 |
| R300Q | - | 2.91 | −16.612 | 96.36 ± 11.27 |
| Dual-mutation | 6′Thr(A) with Fipronil | 2.21 | −12.616 | 124.75 ± 16.03 |
| No. | ΔEvdw | ΔEele | ΔGPB | ΔGSA | ΔGbinding |
|---|---|---|---|---|---|
| Fipronil-WT | −169.155 | −59.646 | 105.167 | −17.233 | −140.868 |
| Fipronil-A302S | −187.221 | −127.879 | 171.830 | −18.084 | −161.355 |
| Fipronil-R300Q | −214.681 | −50.407 | 165.273 | −18.915 | −118.729 |
| Fipronil-Dual | −174.414 | 1.745 | 146.088 | −18.659 | −45.240 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Gao, Y.; Chen, Y.; Liu, G.; Ju, X. Identification of The Fipronil Resistance Associated Mutations in Nilaparvata lugens GABA Receptors by Molecular Modeling. Molecules 2019, 24, 4116. https://doi.org/10.3390/molecules24224116
Tian Y, Gao Y, Chen Y, Liu G, Ju X. Identification of The Fipronil Resistance Associated Mutations in Nilaparvata lugens GABA Receptors by Molecular Modeling. Molecules. 2019; 24(22):4116. https://doi.org/10.3390/molecules24224116
Chicago/Turabian StyleTian, Yafeng, Ya Gao, Yanming Chen, Genyan Liu, and Xiulian Ju. 2019. "Identification of The Fipronil Resistance Associated Mutations in Nilaparvata lugens GABA Receptors by Molecular Modeling" Molecules 24, no. 22: 4116. https://doi.org/10.3390/molecules24224116





