Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics
Abstract
:1. Introduction
2. Results
2.1. Diffuse Reflectance Spectra of Rice Grains
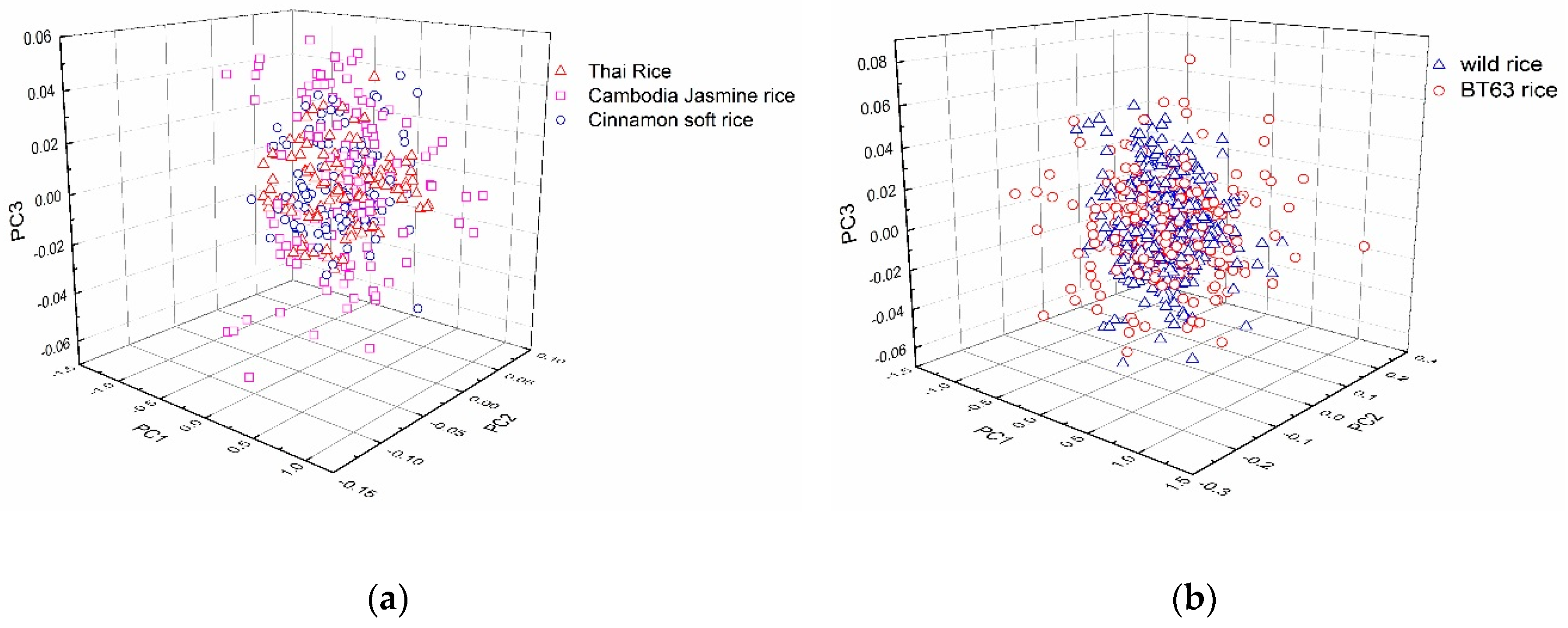
2.2. PCA of Rice Spectra
2.3. PLS-DA Model Establishment and Analysis
2.4. SVM Model Establishment and Analysis
2.5. Selection of Optimal Discriminant Model
3. Discussion
4. Material and Methods
4.1. Rice Samples
4.2. Spectral Measurements
4.3. Spectral Data Pretreatment
4.4. Modeling Methods
4.5. Model Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- James, C. Global Status of Commercialized Biotech/GM Crops: 2014. Available online: https://www.isaaa.org/resources/publications/briefs/49/executivesummary/default.asp (accessed on 13 December 2019).
- Chavas, J.P.; Shi, G.M.; Lauer, J. The effects of gm technology on maize yield. Crop Sci. 2014, 54, 1331–1335. [Google Scholar] [CrossRef] [Green Version]
- Uzogara, S.G. The impact of genetic modification of human foods in the 21st century: A review. Biotechnol. Adv. 2000, 18, 179–206. [Google Scholar] [CrossRef]
- Wang, F.; Ning, D.; Chen, Y.; Dang, C.; Han, N.S.; Liu, Y.E.; Ye, G.Y. Comparing gene expression profiles between BT and non-BT rice in response to brown planthopper infestation. Front. Plant Sci. 2015, 6, 1181. [Google Scholar] [CrossRef] [Green Version]
- Margarit, E.; Reggiardo, M.I.; Vallejos, R.H.; Permingeat, H.R. Detection of BT transgenic maize in foodstuffs. Food Res. Int. 2006, 39, 250–255. [Google Scholar] [CrossRef]
- Mäde, D.; Degner, C.; Grohmann, L. Detection of genetically modified rice: A construct-specific real-time PCR method based on DNA sequences from transgenic BT rice. Eur. Food Res. Technol. 2006, 224, 271–278. [Google Scholar] [CrossRef]
- Akiyama, H.; Sasaki, N.; Sakata, K.; Ohmori, K.; Toyota, A.; Kikuchi, Y.; Watanabe, T.; Furui, S.; Kitta, K.; Maitani, T. Indicated detection of two unapproved transgenic rice lines contaminating vermicelli products. J. Agric. Food Chem. 2007, 55, 5942–5947. [Google Scholar] [CrossRef]
- Grohmann, L.; Mäde, D. Detection of genetically modified rice: Collaborative validation study of a construct-specific real-time PCR method for detection of transgenic BT rice. Eur. Food Res. Technol. 2009, 228, 497–500. [Google Scholar] [CrossRef]
- Singh, C.K.; Ojha, A.; Kachru, D.N. Detection and characterization of cry1Ac transgene construct in BT cotton: Multiple polymerase chain reaction approach. J. AOAC Int. 2007, 90, 1517–1725. [Google Scholar]
- Stull, D. A feat of fluorescence. Scientist 2001, 15, 20–21. [Google Scholar]
- Duijn, G.J.; Biert, R.; Bleeker-Marcelis, H.; Boeijen, I.; Adan, A.J.; Jhakrie, S.; Hessing, M. Detection of genetically modified organisms in foods by protein- and DNA-based techniques: Bridging the methods. J. AOAC Int. 2002, 85, 787–791. [Google Scholar] [PubMed]
- Alishahi, A.; Farahmand, H.; Prieto, N.; Cozzolino, D. Identification of transgenic foods using NIR spectroscopy: A review. Spectrochim. Acta Part. A 2010, 75, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.J.; Ying, Y.B.; Ying, T.J.; Yu, H.Y.; Fu, X.P. Distrimination of transgenic tomatoes based on visible/near-infrared spectra. Anal. Chim. Acta 2007, 584, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.S.; da Silva, A.P.; Pinho, J.S.; Ferré, J.; Boqué, R. Rapid characterization of transgenic and non-transgenic soybean oils by chemometric methods using NIR spectroscopy. Spectrochim. Acta Part. A 2013, 100, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Kleinbaum, D.; Kupper, L.; Muller, K. Applied Regression Analysis and Other Multivariate Methods, 2nd ed.; Duxbury Press: Belmont, CA, USA, 1988; pp. 657–662. [Google Scholar]
- Zhang, H.Y.; Ding, D.; Song, L.Q.; Gu, L.N.; Tang, Y.G. Achievement of the noninvasive measurement for human blood glucose with NIR diffusion reflectance spectrum method. Spectrocopy Spectr. Anal. 2005, 6, 882–885. [Google Scholar]
- Huang, L.S.; Liu, D.; Qin, T.J.; Lin, X.W.; Zhou, Z.H.; Huang, J.L.; Cong, Y.L. Near infrared spectral quantitative model of the nutrient content in rice. Mod. Food Sci. Technol. 2019, 35, 317–324. [Google Scholar]
- Teye, E.; Amuah, C.L.Y.; McGrath, T.; Elliott, C. Innovative and rapid analysis for rice authenticity using hand-held NIR spectrometry and chemometrics. Spectrochim. Acta Part A 2019, 217, 147–154. [Google Scholar] [CrossRef]
- Xu, L.; Yan, S.M.; Cai, C.B.; Yu, X.P. Untargeted detection of Illegal adulterations in Chinese glutinous rice flour (GRF) by NIR spectroscopy and chemometrics: Specificity of detection improved by reducing unnecessary variations. Food Anal. Methods 2013, 6, 1568–1575. [Google Scholar] [CrossRef]
- Sanchis, V.; Bourguet, D. Bacillus thuringiensis: Applications in agriculture and insect resistance management. A review. Agron. Sustain. Dev. 2008, 28, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.X.; Hu, W.; Zhou, Y.C.; Zhang, X.Y.; Xu, S.; Guo, Q.Y.; Qi, P.; Chen, L.L.; Yang, X.Z.; Zhang, F.; et al. Use of near-infrared spectroscopy for the rapid evaluation of soybean [Glycine max (L.) Merri.] water soluble protein content. Spectrochim. Acta. Part. A 2020, 224, 117400. [Google Scholar] [CrossRef]
- Wiedemair, V.; Mair, D.; Held, C.; Huck, C.W. Investigations into the use of handheld near-infrared spectrometer and novel semi-automated data analysis for the determination of protein content in different cultivars of Panicum miliaceumL. Talanta 2019, 205, 120115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lu, J.G. Using an optimal CC-PLSR-RBFNN model and NIR spectroscopy for the starch content determination in corn. Spectrochim. Acta. Part. A 2018, 196, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Y.Y.; Peng, Y.K.; Yang, B.N.; Li, L.; Liu, Y.C. Multi-parameter potato quality non-destructive rapid detection by visible/near-infrared spectra. Spectrosc. Spectra Anal. 2018, 38, 3736–3742. [Google Scholar]
- Shao, Y.; Cen, Y.; He, Y.; Liu, F. Infrared spectroscopy and chemometrics for the starch and protein prediction in irradiated rice. Food Chem. 2011, 126, 1856–1861. [Google Scholar] [CrossRef]
- Xavier, H.; Nathalie, G.; Roger, J.M.; Ryad, B.; Gilles, R. Comparison of the efficacy of spectral pre-treatments for wheat and weed discrimination in outdoor conditions. Comput. Electron. Agric. 2014, 108, 242–249. [Google Scholar]
- Esquerre, C.; Gowen, A.A.; Burger, J.; Downey, G.; O’Donnell, C.P. Suppressing sample morphology effects in near infrared spectral imaging using chemometric data pre-treatments. Chemom. Intell. Lab. Syst. 2012, 117, 129–137. [Google Scholar] [CrossRef]
- Palou, A.; Miró, A.; Blanco, M.; Larraz, R.; Gómez, J.F.; Martínez, T.; González, J.M.; Alcalà, M. Calibration sets selection strategy for the construction of robust PLS models for prediction of biodiesel/diesel blends physico-chemical properties using NIR spectroscopy. Spectrochim. Acta Part. A 2017, 180, 119–126. [Google Scholar] [CrossRef]
- Xie, L.J.; Ying, Y.B.; Ying, T.J. Combination and comparison of chemometrics methods for identification of transgenic tomatoes using visible and near-infrared diffuse transmittance technique. J. Food Eng. 2007, 82, 395–401. [Google Scholar] [CrossRef]
- Chen, Y. Reference-related component analysis: A new method inheriting the advantages of PLS and PCA for separating interesting information and reducing data dimension. Chemom. Intell. Lab. Syst. 2016, 156, 196–202. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2011, 58, 109–130. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Soares, A.; Castanho, A.; Almeida, A.S.; Oliveira, J.; Brites, C. Optimization of rice amylose determination by NIR-spectroscopy using PLS chemometrics algorithms. Food Chem. 2018, 242, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Marengo, E.; Robotti, E.; Bobba, M.; Milli, A.; Campostrini, N.; Righetti, S.C.; Cecconi, D.; Righetti, P.G. Application of partial least squares discriminant analysis and variable selection procedures: A 2D-PAGE proteomic study. Anal. Bioanal. Chem. 2008, 390, 1327–1342. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Van Gestel, T.; Suykens, J.A.; Baesens, B.; Viaene, S.; Vanthienen, J.; Dedene, G.; Vandewalle, J. Benchmarking least squares support vector machine classifiers. Mach. Learn. 2004, 54, 5–32. [Google Scholar] [CrossRef]
- Xu, J.C.; Ren, Q.W.; Shen, Z.Z. Prediction of the strength of concrete radiation shielding based on LS-SVM. Ann. Nucl. Energy 2015, 85, 296–300. [Google Scholar]
- Julio, C.L.; Ronei, J.P. Biodiesel content determination in diesel fuel blends using near infrared (NIR) spectroscopy and support vector machines (SVM). Talanta 2013, 104, 155–161. [Google Scholar]
- Wang, X.K.; Guan, S.Y.; Hua, L.; Wang, B.; He, X.M. Classification of spot-welded joint strength using ultrasonic signal time-frequency features and PSO-SVM method. Ultrasonics 2019, 91, 161–169. [Google Scholar] [CrossRef]
- Galan, S.G.; Prado, R.P.; Exposito, J.E.M. Rules discovery in fuzzy classifier systems with PSO for scheduling in grid computational infrastructures. Appl. Soft Comput. 2015, 29, 424–435. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |




| Methods | No. of LVs 1 | Accuracy (%) |
|---|---|---|
| Origin | 8 | 95.83 |
| NWS 2 | 14 | 97.50 |
| SNV 2 | 7 | 95.00 |
| MSC 2 | 7 | 95.00 |
| SG 1st-Der 2 | 11 | 95.83 |
| Methods | No. of LVs | Accuracy (%) |
|---|---|---|
| Origin | 14 | 98.75 |
| NWS | 20 | 99.17 |
| SNV | 16 | 100.00 |
| MSC | 16 | 100.00 |
| SG 1st-Der | 14 | 100.00 |
| Methods | C 1/Gamma | Accuracy (%) |
|---|---|---|
| Origin | 92.70/3.49 | 92.70 |
| NWS | 34.73/16.78 | 93.33 |
| SNV | 7.81/26.08 | 98.33 |
| MSC | 7.00/969.54 | 98.75 |
| SG 1st-Der | 93.34/1000 | 99.58 |
| Methods | C/Gamma | Accuracy (%) |
|---|---|---|
| Origin | 100/10.9 | 99.38 |
| NWS | 100/14.33 | 99.79 |
| SNV | 18.63/35.68 | 100.00 |
| MSC | 17.42/1000 | 100.00 |
| SG 1st-Der | 78.02/1000 | 99.58 |
| Types | Number of Samples |
|---|---|
| Cambodia Jasmine rice | 120 |
| Thai rice | 120 |
| Cinnamon soft rice | 120 |
| Bt-SY63 rice | 360 |
| SY63 rice | 360 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Geng, P.; Wu, W.; Wen, Q.; Rao, M. Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics. Molecules 2019, 24, 4568. https://doi.org/10.3390/molecules24244568
Hao Y, Geng P, Wu W, Wen Q, Rao M. Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics. Molecules. 2019; 24(24):4568. https://doi.org/10.3390/molecules24244568
Chicago/Turabian StyleHao, Yong, Pei Geng, Wenhui Wu, Qinhua Wen, and Min Rao. 2019. "Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics" Molecules 24, no. 24: 4568. https://doi.org/10.3390/molecules24244568
APA StyleHao, Y., Geng, P., Wu, W., Wen, Q., & Rao, M. (2019). Identification of Rice Varieties and Transgenic Characteristics Based on Near-Infrared Diffuse Reflectance Spectroscopy and Chemometrics. Molecules, 24(24), 4568. https://doi.org/10.3390/molecules24244568






