Cyclic Peptide-Based Sirtuin Substrates
Abstract
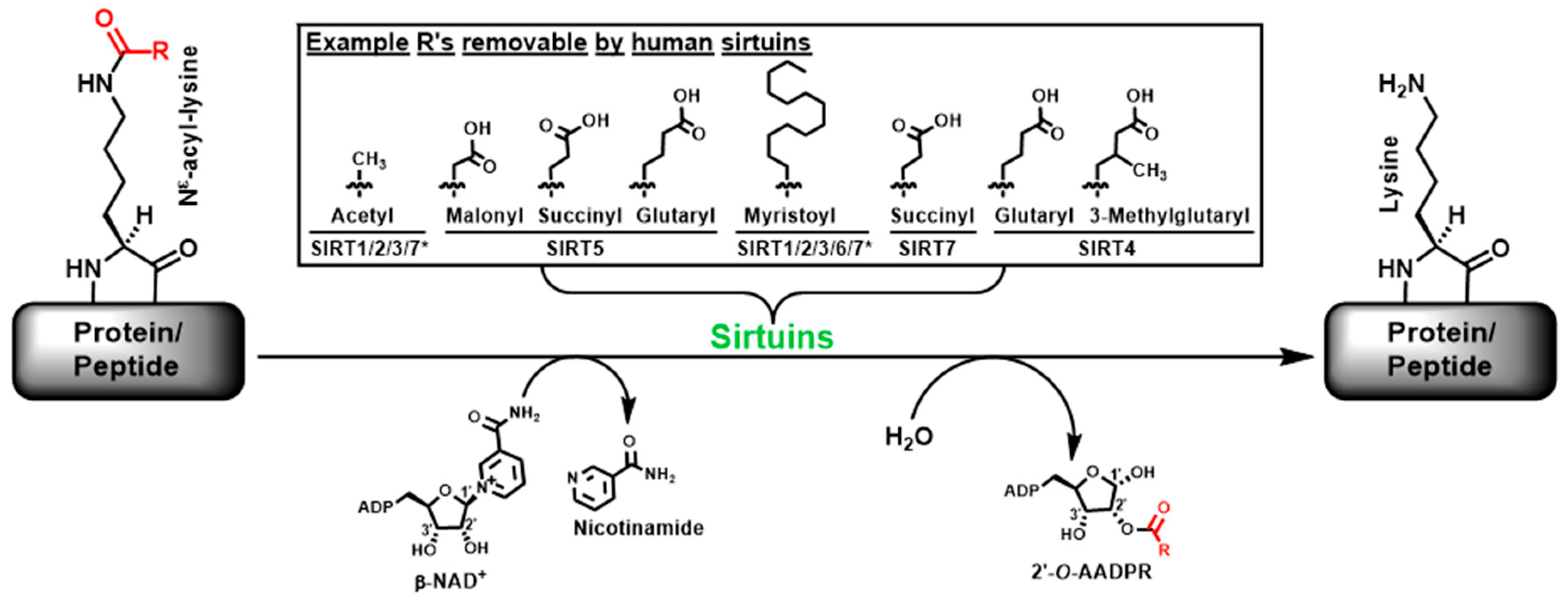
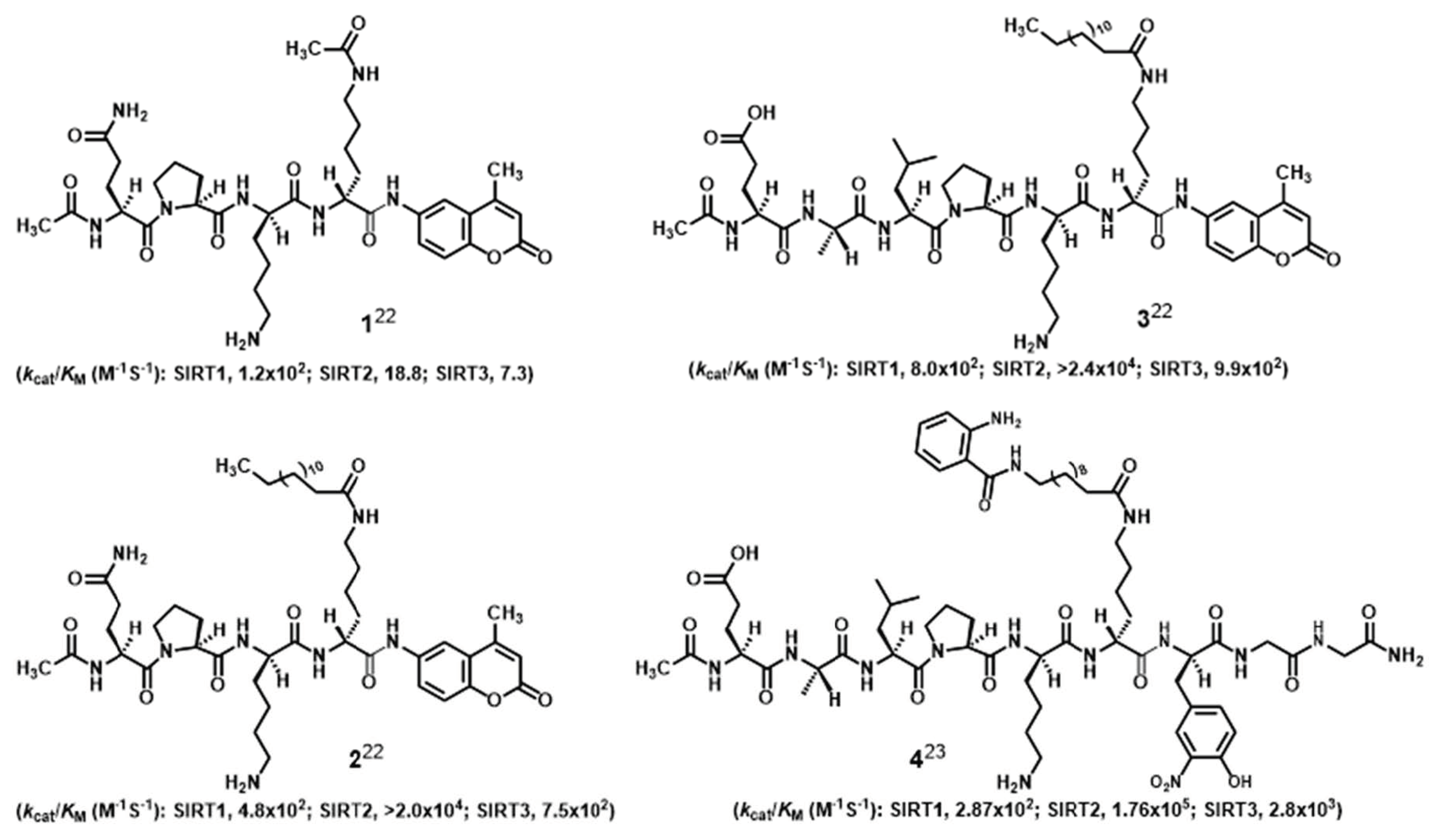
:1. Introduction
2. Results and Discussion
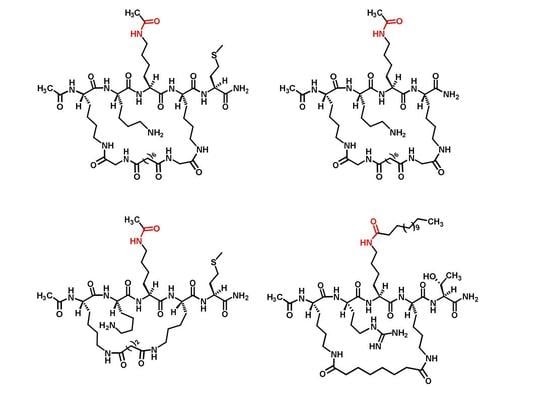
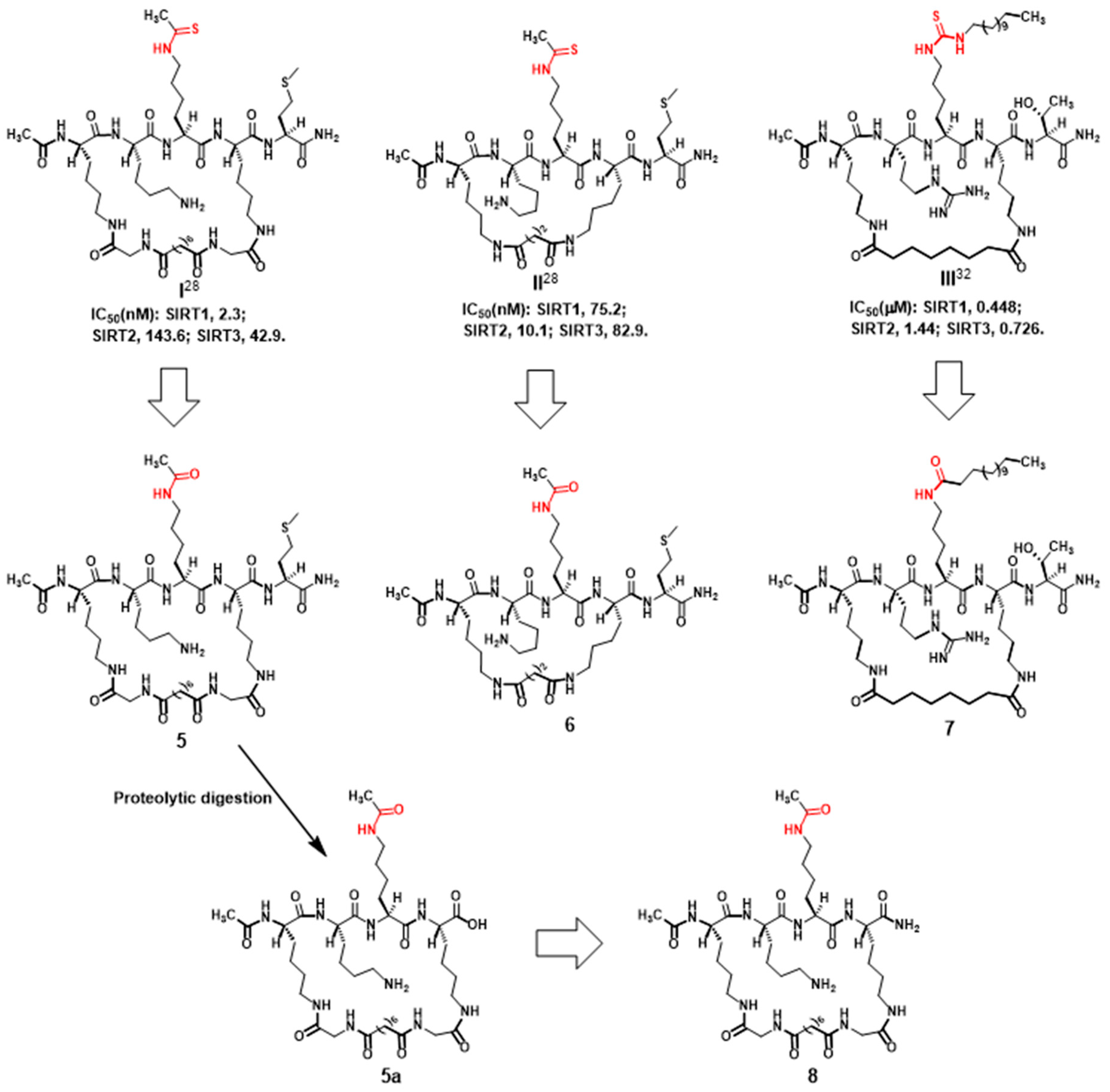
2.1. Compound Design
2.2. Compound Preparation
2.3. Compound Evaluation with Pronase Digestion Assay
3. Experimental Section
3.1. General
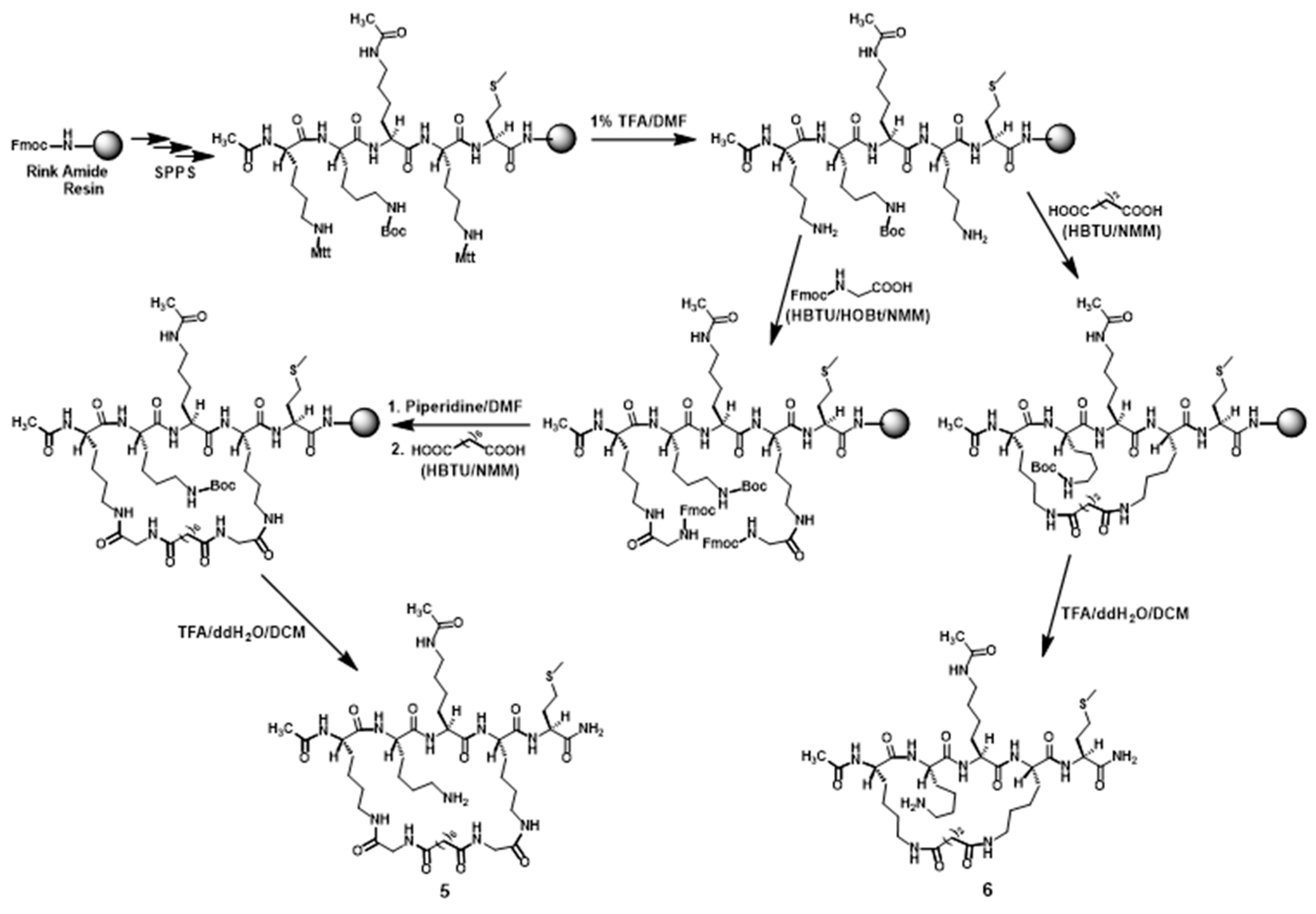
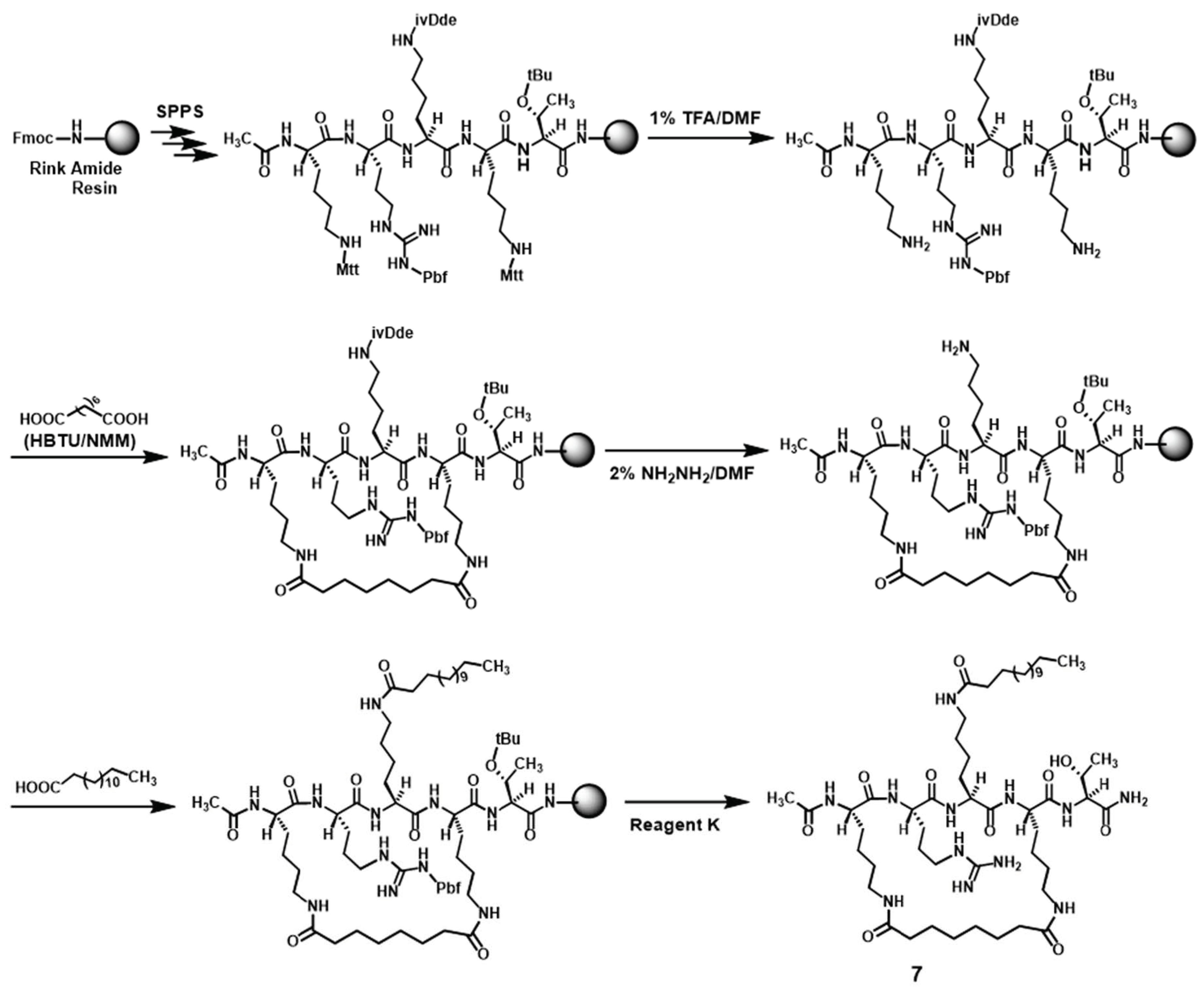
3.2. Compound Preparation
3.2.1. Preparation of Compounds 5 and 6 (Scheme 1)
3.2.2. Preparation of Compound 7 (Scheme 2)
3.2.3. Preparation of Compound 8 (Scheme 3)
3.3. Kinetic Parameter Determination for 5–8’s Substrate Activities with SIRT1/2/3
3.4. Pronase Digestion Assay
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greiss, S.; Gartner, A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 2009, 28, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhou, Y.; Wang, F.; Chen, X.; Wang, C.; Wang, J.; Liu, T.; Li, Y.; He, B. SIRT4 is the last puzzle of mitochondrial sirtuins. Bioorg. Med. Chem. 2018, 26, 3861–3865. [Google Scholar] [PubMed]
- Hu, X.; Zheng, W. Chemical probes in sirtuin research. In Progress in Molecular Biology and Translational Science. Sirtuins in Health and Disease, 1st ed.; Zheng, W., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 154, pp. 1–24. [Google Scholar]
- Rajabi, N.; Galleano, I.; Madsen, A.S.; Olsen, C.A. Targeting sirtuins: Substrate specificity and inhibitor design. In Progress in Molecular Biology and Translational Science. Sirtuins in Health and Disease, 1st ed.; Zheng, W., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 154, pp. 25–69. [Google Scholar]
- Li, S.; Zheng, W. Mammalian sirtuins SIRT4 and SIRT7. In Progress in Molecular Biology and Translational Science. Sirtuins in Health and Disease, 1st ed.; Zheng, W., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 154, pp. 147–168. [Google Scholar]
- Bheda, P.; Jing, H.; Wolberger, C.; Lin, H. The substrate specificity of sirtuins. Annu. Rev. Biochem. 2016, 85, 405–429. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zang, W.; Wang, J.; Huang, Y.; He, Y.; Yan, L.; Liu, J.; Zheng, W. The chemical biology of sirtuins. Chem. Soc. Rev. 2015, 44, 5246–5264. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Redondo, P.; Vaquero, A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer 2013, 4, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Lombard, D.B. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Elkhwanky, M.S.; Hakkola, J. Extranuclear sirtuins and metabolic stress. Antioxid. Redox. Signal. 2018, 28, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Bringman-Rodenbarger, L.R.; Guo, A.H.; Lyssiotis, C.A.; Lombard, D.B. Emerging roles for SIRT5 in metabolism and cancer. Antioxid. Redox. Signal. 2018, 28, 677–690. [Google Scholar] [CrossRef]
- Sebastián, C.; Mostoslavsky, R. The role of mammalian sirtuins in cancer metabolism. Semin. Cell Dev. Biol. 2015, 43, 33–42. [Google Scholar] [CrossRef]
- Choi, J.E.; Mostoslavsky, R. Sirtuins, metabolism, and DNA repair. Curr. Opin. Genet. Dev. 2014, 26, 24–32. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Faller, D.V. Transcription regulation by class III histone deacetylases (HDACs)-sirtuins. Transl. Oncogenomics 2008, 3, 53–65. [Google Scholar] [PubMed]
- Wang, Y.; He, J.; Liao, M.; Hu, M.; Li, W.; Ouyang, H.; Wang, X.; Ye, T.; Zhang, Y.; Ouyang, L. An overview of sirtuins as potential therapeutic target: Structure, function and modulators. Eur. J. Med. Chem. 2019, 161, 48–77. [Google Scholar] [CrossRef] [PubMed]
- Neo, S.H.; Tang, B.L. Sirtuins as modifiers of huntington’s disease (HD) pathology. In Progress in Molecular Biology and Translational Science. Sirtuins in Health and Disease, 1st ed.; Zheng, W., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 154, pp. 105–145. [Google Scholar]
- Schiedel, M.; Robaa, D.; Rumpf, T.; Sippl, W.; Jung, M. The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med. Res. Rev. 2018, 38, 147–200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, J.; Chen, D.; Yan, L.; Zheng, W. Sirtuin inhibition: Strategies, inhibitors, and therapeutic potential. Trends Pharmacol. Sci. 2017, 38, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Zhang, X.; Lin, H. HPLC-based enzyme assays for sirtuins. Methods Mol. Biol. 2018, 1813, 225–234. [Google Scholar] [PubMed]
- Galleano, I.; Schiedel, M.; Jung, M.; Madsen, A.S.; Olsen, C.A. A continuous, fluorogenic sirtuin 2 deacylase assay: Substrate screening and inhibitor evaluation. J. Med. Chem. 2016, 59, 1021–1031. [Google Scholar] [CrossRef]
- Chiang, Y.L.; Lin, H. An improved fluorogenic assay for SIRT1, SIRT2, and SIRT3. Org. Biomol. Chem. 2016, 14, 2186–2190. [Google Scholar] [CrossRef] [Green Version]
- Schuster, S.; Roessler, C.; Meleshin, M.; Zimmermann, P.; Simic, Z.; Kambach, C.; Schiene-Fischer, C.; Steegborn, C.; Hottiger, M.O.; Schutkowski, M. A continuous sirtuin activity assay without any coupling to enzymatic or chemical reactions. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Li, Y.; Liu, T.; Liao, S.; Li, Y.; Lan, Y.; Wang, A.; Wang, Y.; He, B. A mini-review on sirtuin activity assays. Biochem. Biophys. Res. Commun. 2015, 467, 459–466. [Google Scholar] [CrossRef]
- Roessler, C.; Tüting, C.; Meleshin, M.; Steegborn, C.; Schutkowski, M. A novel continuous assay for the deacylase sirtuin 5 and other deacetylases. J. Med. Chem. 2015, 58, 7217–7223. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.B.; Jing, H.; Aramsangtienchai, P.; He, B.; Khan, S.; Hu, J.; Lin, H.; Hao, Q. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Goodman, M.; Ro, S. Peptidomimetics for drug design. In Burger’s Medicinal Chemistry and Drug Discovery. Principles and Practice, 5th ed.; Wolff, M.E., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 1995; Volume 1, pp. 803–861. [Google Scholar]
- Huang, Y.; Liu, J.; Yan, L.; Zheng, W. Simple Nε-thioacetyl-lysine-containing cyclic peptides exhibiting highly potent sirtuin inhibition. Bioorg. Med. Chem. Lett. 2016, 26, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Fatkins, D.G.; Monnot, A.D.; Zheng, W. Nε-thioacetyl-lysine: A multi-facet functional probe for enzymatic protein lysine Nε-deacetylation. Bioorg. Med. Chem. Lett. 2006, 16, 3651–3656. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Denu, J.M. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry 2007, 46, 14478–14486. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zheng, W. Cyclic peptide-based potent human SIRT6 inhibitors. Org. Biomol. Chem. 2016, 14, 5928–5935. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, B.M.; Hao, Y.; Li, X.; Wesdemiotis, C.; Wang, Z.; Zheng, W. A mechanism-based potent sirtuin inhibitor containing Nε-thiocarbamoyl-lysine (TuAcK). Bioorg. Med. Chem. Lett. 2011, 21, 4753–4757. [Google Scholar] [CrossRef] [PubMed]
- Roche Applied Science. Pronase: Product Description. Available online: https://www.sigmaaldrich.com/catalog/product/roche/pronro?lang=en®ion=SX (accessed on 19 January 2019).
- Hirsch, B.M.; Gallo, C.A.; Du, Z.; Wang, Z.; Zheng, W. Discovery of potent, proteolytically stable, and cell permeable human sirtuin peptidomimetic inhibitors containing Nε-thioacetyl-lysine. Med. Chem. Comm. 2010, 1, 233–238. [Google Scholar] [CrossRef]


| Compound | Ionic Formula | Calculated m/z | Observed m/z |
|---|---|---|---|
| 5 | [C45H81N12O11S]+ | 997.5863 | 997.5856 |
| 6 | [C37H67N10O9S]+ | 827.4808 | 827.4778 |
| 7 | [C52H97N12O10]+ | 1049.7445 | 1049.7449 |
| 8 | [C40H71N11O10Na]+ | 888.5278 | 888.5282 |
| Compound | 5 | 6 | ||||
|---|---|---|---|---|---|---|
| Sirtuin | KM (μM) | kcat (10−3, s−1) | kcat/KM (M−1·s−1) | KM (μM) | kcat (10−3, s−1) | kcat/KM (M−1·s−1) |
| SIRT1 | 22.2 ± 1.8 | 35.6 ± 5.1 | (1.6 ± 0.1) × 103 | 41.3 ± 1.83 | 25.9 ± 2.3 | (0.63 ± 0.03) × 103 |
| SIRT2 | 11.6 ± 2.2 | 36.0 ± 4.9 | (3.2 ± 1.0) × 103 | 49.7 ± 7.78 | 138.9 ± 26.7 | (2.79 ± 0.1) × 103 |
| SIRT3 | 4.5 ± 0.9 | 15.8 ± 1.4 | (3.6 ± 0.4) × 103 | 34.9 ± 4.04 | 155.3 ± 16.6 | (4.51 ± 1.0) × 103 |
| Compound | 7 | 8 | ||||
|---|---|---|---|---|---|---|
| Sirtuin | KM (μM) | kcat (10−3, s−1) | kcat/KM (M−1·s−1) | KM (μM) | kcat (10−3, s−1) | kcat/KM (M−1·s−1) |
| SIRT1 | 2.85 ± 0.13 | 5.8 ± 0.1 | (2.04 ± 0.06) × 103 | 42.2 ± 17.5 | 90.2 ± 39.4 | (2.2 ± 0.05) × 103 |
| SIRT2 | 37.6 ± 14.4 | 90.9 ± 2.62 | (2.6 ± 0.93) × 103 | 43.1 ± 16.0 | 414.0 ± 25.0 | (10.4 ± 4.4) × 103 |
| SIRT3 | 4.2 ± 0.38 | 60.0 ± 2.8 | (14.3 ± 0.66) × 103 | 99.9 ± 48.7 | 221.0 ± 32.0 | (2.4 ± 0.9) × 103 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Yan, L.; Zheng, W. Cyclic Peptide-Based Sirtuin Substrates. Molecules 2019, 24, 424. https://doi.org/10.3390/molecules24030424
Chen D, Yan L, Zheng W. Cyclic Peptide-Based Sirtuin Substrates. Molecules. 2019; 24(3):424. https://doi.org/10.3390/molecules24030424
Chicago/Turabian StyleChen, Di, Lingling Yan, and Weiping Zheng. 2019. "Cyclic Peptide-Based Sirtuin Substrates" Molecules 24, no. 3: 424. https://doi.org/10.3390/molecules24030424