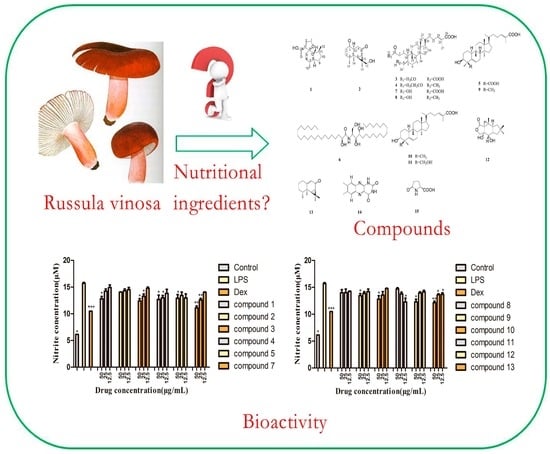
Chemical Constituents with Inhibitory Activity of NO Production from a Wild Edible Mushroom, Russula vinosa Lindbl, May Be Its Nutritional Ingredients
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structural Elucidation
2.2. Marked Peaks of Isolated Compounds
2.3. Bioactivity Evaluation
2.3.1. Cytotoxic Activity Assay
2.3.2. Inhibitory Activity on NO Production Assay
3. Materials and Methods
3.1. General
3.2. UPLC-QTOF-MS/MS Conditions
3.3. Plant Materials
3.4. Extraction and Isolation
3.4.1. Vinosane (1)
3.4.2. Rulepidadione C (2)
3.4.3. (24E)-3,4-Seco-cucurbita-4,24-diene-26,29-dioic acid-3-methyl ester (3)
3.4.4. (24E)-3,4-Seco-cucurbita-4,24-diene-26-oic acid-3-ethyl ester (4)
3.4.5. (24E)-3β-Hydroxycucurbita-5,24-diene-26,29-dioic acid (5)
3.4.6. (2S,3S,4R,2′R)-2-(2′-Hydroxydocosanoylamino) eicosane-1,3,4-triol (6)
3.5. Biological Activity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, H.; Huang, D.; Xu, X.; Zhang, D.; Shi, Q.; Wu, S. Studies on the chemical compositions in Russula vinosa. Mycosystema 1998, 17, 68–74. [Google Scholar]
- Song, B.; Li, T.; Wu, X.; Li, J.; Shen, Y.; Lin, Q. Known species of Russula from China and their distribution. J. Fung. Res. 2007, 5, 20–42. [Google Scholar]
- Zhang, Y.; Xia, A.; Liang, Y.; Kang, Z.; Zhou, N.; Zhang, Q. Research progress on pharmacological action of Russula. China Pharm. 2013, 22, 95–96. [Google Scholar]
- Yang, Y.-L.; Ren, J.-L.; Zhang, H. Research progress of terpenoids and bioactivities in edible mushroom. Sci. Tech. Food Indust. 2019, 40, 309–310. [Google Scholar]
- Ou, Y.X.; Li, Y.Y.; Qian, X.M.; Shen, Y.M. Guanacastane-type diterpenoids from Coprinus radians. Phytochemistry 2012, 78, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Boonsong, S.; Klaypradit, W.; Wilaipun, P. Antioxidant activities of extracts from five edible mushrooms using different extactants. Agric. Nat. Res. 2016, 50, 89–97. [Google Scholar]
- Yun, B.S.; Lee, I.K.; Cho, Y.; Cho, S.M.; Yoo, I.D. New tricyclic sesquiterpenes from the fermentation broth of Stereum hirsutum. J. Nat. Prod. 2002, 65, 786–788. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Matsumoto, Y.; Hama, H.; Tanaka, M.; Zhai, H.; Fukuyama, Y.; Arihara, S.; Hashimoto, T. Russujaponols G-L, illudoid sesquiterpenes and their neurite outgrowth promoting activity from the fuit body of Russula japonica. Chem. Pharm. Bull. 2009, 57, 311–314. [Google Scholar] [CrossRef]
- Matsuura, M.; Kato, S.; Saikawa, Y.; Nakata, M.; Hashimoto. Identification of Cyclopropylacetyl-R -carnitine, a unique chemical marker of the fatally toxic mushroom Russula subnigricans. Chem. Pharm. Bull. 2016, 64, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Maarisit, W.; Abdjul, D.B.; Tamazaki, H.; Takahashi, O.; Kirikoshi, R.; Kanno, S.; Namikoshi, M. Structures and biological activities of triterpenes and sesquiterpenes obtained from Russula lepida. Phytochemistry 2016, 127, 63–68. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Shen, X.; Chao, X.; Ho, C.C.; Cheng, X.L.; Zhang, Y.; Lin, R.C.; Du, K.J.; Luo, W.-J.; Chen, J.-Y.; et al. Ergosta-4,6,8(14)-tetraen-3-one induce G2/M cell cycle arrest and apotosis in human hepatocellular carcinoma cells. Biochim. Biophys. Acta 2011, 1810, 384–390. [Google Scholar] [CrossRef]
- Daniewski, W.M.; Gumulka, M.; Ptaszynska, K.; Skibicki, P.; Krajewski, J.; Gluzinski, P. Marasmane lactones from Lactarius vellereus. Phytochemistry 1992, 31, 913–915. [Google Scholar] [CrossRef]
- Hao, X.; Tan, N.; Zhou, J. Constituents of Castrodia elata in Guizhou. Acta Botanica Yunnanica 2000, 22, 81–84. [Google Scholar]
- Kwon, H.C.; Kim, K.R.; Zee, S.D.; Cho, S.Y.; Lee, K.R. A new indolinepeptide from paecilomyces sp. J300. Arch. Pharm. Res. 2004, 27, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Shi, D.Y.; Li, J.; Guo, S.J.; Li, L.L.; Yuan, Z.H.; Zhu, X.B. Sesquiterpenes from Laurencia similis. Molecules 2009, 14, 1889–1897. [Google Scholar] [CrossRef]
- Tan, J.-W.; Dong, Z.-J.; Liu, J.-K. New terpenoids from basidiomycetes Russula lepida. Helv. Chim. Acta 2000, 83, 3191–3197. [Google Scholar]
- Wang, H.; Yang, G.; Wu, S.; Wang, S.; Li, G.; Li, W.; Meng, L.; Li, Z. Studies on the chemical contituent of Russula rosacea. Acta Pharm. Sin. 1994, 29, 39–43. [Google Scholar]
- Qin, B.H.; Liu, X.Q.; Yuan, Q.Y.; Wang, J.; Han, H.Y. Anti-Inflammatory triterpenoids from the Caulophyllum robustum maximin LPS-stimulated RAW 264.7 Cells. Molecules 2018, 23, 1149. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.M.; Zhang, L.C.; Liu, J.K. Paxillamide: a novel phytosphingosine derivative from the fruiting bodies of Paxillus panuoides. Helv. Chim. Acta 2004, 87, 1483–1487. [Google Scholar] [CrossRef]
- Lun, B.S.; Shao, L.W.; Wang, Y.; Li, C.C.; Yu, H.; Wang, C.H.; Zhu, Y. Taxanes from Taxus wallichiana var. mairei cultivated in the southern area of the Yangtze River in China. Nat. Prod. Res. 2017, 31, 2341–2347. [Google Scholar] [CrossRef]
- Xiang, W.; Zhang, G.D.; Li, F.Y.; Wang, T.L.; Suo, T.C.; Wang, C.H.; Li, Z.; Zhu, Y. Chemical constituents from the roots of Polygala arillata and their anti-inflammatory activities, J. Chem. 2019, 2019, 8079619. [Google Scholar] [CrossRef]
- Chen, X.-H.; Xia, L.-X.; Zhou, H.-B.; Qiu, G.-Z. Chemical composition and antioxidant activities of Russula griseocarnosa sp. nov. J. Agric. Food Chem. 2010, 58, 6966–6971. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |






| No. | 1 a | 2 b | 3 a | 4 a | 5 c |
|---|---|---|---|---|---|
| 1 | 4.78 (m) | 1.68 (m), 1.95 (m) | 1.26 (m), 1.85 (m) | 1.60 (m) | |
| 2 | 6.82 (s) | 2.33 (m), 2.58 (m) | 2.31 (m) | 2.25 (m) | 2.03 (m) |
| 3 | 1.75 (m), 2.02 (m) | 4.23 (m) | |||
| 4 | 2.27 (m) | ||||
| 5 | 1.56 (m), 1.34 (m) | ||||
| 6 | 1.28 (m), 1.53 (m) | 1.49 (d, 8.1) | 2.14 (m), 3.13 (m) | 2.00 (m), 2.42 (m) | 6.01 (d,5.6) |
| 7 | 1.75 (m) | 1.75 (m) | 1.24 (m), 1.85 (m) | 1.39 (m), 1.93 m) | 2.31 (m), 1.75 (m) |
| 8 | 2.05 (m) | 2.48 (dd, 7.5, 5.7) | 1.93 (m) | 1.67 (m) | |
| 9 | 1.61 (m), 1.48 (m) | 2.16 (m), 2.49 (m) | |||
| 10 | 1.48 (m) | 3.08 (dd, 12.6, 5.2) | 1.96 (1H, m) | 2.42 (m) | 2.44 (m) |
| 11 | 2.12 (m) | 1.50 (m) | 1.58 (m), 1.71 (m) | 1.67 (m), 1.39 (m) | |
| 12 | 1.86 (t, 6.0) | 3.37 (m) | 1.25 (m), 1.50 (m) | 1.12 (m) | 1.47 (m) |
| 13 | 0.96 (d, 7.2) | 1.42 (s) | |||
| 14 | 0.88 (s) | ||||
| 15 | 0.90 (d, 6.8) | 1.15 (d, 6.8) | 1.13 (m) | 1.49 (m), 1.70 (m) | 1.61 (m), 1.45 (m) |
| 16 | 1.50 (m), 1.98 (m) | 1.56 (m), 1.86 (m) | 1.84 (m), 1.20 (m) | ||
| 17 | 1.49 (m) | 1.52 (m) | 1.47 (m) | ||
| 18 | 0.82 (s) | 0.81 (s) | 0.81 (s) | ||
| 19 | 1.19 (s) | 1.14 (s) | 0.97 (s) | ||
| 20 | 1.44 (m) | 1.46 (m) | 1.48 (m) | ||
| 21 | 0.91 (d, 6.0) | 0.91 (d,6.0) | 0.97 (s) | ||
| 22 | 1.50 (m) | 1.19 (m), 1.55 (m) | 1.05 (m), 1.15 (m) | ||
| 23 | 2.24 (m), 2.09 (m) | 2.12 (m), 2.26 (m) | 2.39 (m), 2.15 (m) | ||
| 24 | 6.90 (m) | 6.90 (s) | 7.23 (m) | ||
| 25 | |||||
| 26 | |||||
| 27 | 1.84 (s) | 1.83 (s) | 2.11 (s) | ||
| 28 | 1.84 (s) | 1.64 (s) | 1.66 (s) | ||
| 29 | 1.54 (s) | ||||
| 30 | 0.66 (3H, s) | 0.63 (3H, s) | 0.90 (s) | ||
| OCH3(2) | 3.66 (3H, s) | 4.13 (2H, m) | |||
| CH3 | 1.25 (3H, t, 7.1) |
| No. | 1 a | 2 b | 3 a | 4 a | 5 c |
|---|---|---|---|---|---|
| 1 | 72.2 | 211.8 | 26.2 | 27.9 | 21.5 |
| 2 | 129.5 | 41.5 | 32.5 | 33.1 | 29.6 |
| 3 | 152.3 | 32.0 | 174.4 | 174.7 | 72.9 |
| 4 | 92.6 | 40.5 | 121.9 | 123.2 | 55.0 |
| 5 | 37.3 | 41.0 | 154.7 | 133.8 | 138.8 |
| 6 | 24.1 | 35.4 | 24.6 | 22.4 | 123.3 |
| 7 | 32.4 | 33.8 | 27.9 | 22.5 | 24.9 |
| 8 | 46.1 | 211.2 | 43.8 | 43.9 | 44.2 |
| 9 | 23.4 | 35.2 | 38.1 | 37.5 | 35.4 |
| 10 | 31.8 | 51.4 | 43.6 | 42.8 | 37.7 |
| 11 | 39.3 | 33.8 | 36.9 | 36.8 | 32.7 |
| 12 | 49.0 | 72.0 | 30.0 | 34.4 | 35.9 |
| 13 | 17.2 | 13.8 | 45.8 | 45.9 | 46.9 |
| 14 | 168.0 | 16.0 | 48.9 | 49.2 | 49.9 |
| 15 | 20.1 | 15.5 | 34.2 | 30.3 | 31.1 |
| 16 | 23.0 | 26.6 | 28.5 | ||
| 17 | 51.0 | 51.0 | 51.1 | ||
| 18 | 15.4 | 15.6 | 15.9 | ||
| 19 | 30.7 | 30.8 | 28.2 | ||
| 20 | 36.3 | 36.2 | 36.5 | ||
| 21 | 18.6 | 18.6 | 19.1 | ||
| 22 | 35.1 | 35.0 | 35.2 | ||
| 23 | 26.0 | 26.0 | 26.3 | ||
| 24 | 146.0 | 146.0 | 142.9 | ||
| 25 | 126.8 | 126.7 | 129.3 | ||
| 26 | 173.8 | 173.2 | 171.0 | ||
| 27 | 12.0 | 12.1 | 13.2 | ||
| 28 | 16.5 | 20.4 | 23.1 | ||
| 29 | 175.8 | 21.4 | 180.5 | ||
| 30 | 17.4 | 17.4 | 18.2 | ||
| OCH3(2) | 51.7 | 60.3 | |||
| CH3 | 14.4 |
| Position | 13C | 1H |
|---|---|---|
| 1 | 62.4 | 4.53 (m), 4.38 (m) |
| 2 | 53.4 | 4.45 (dd, 10.8, 4.5) |
| 3 | 77.2 | 5.14 (m) |
| 4 | 73.4 | 4.31 (m) |
| 5 | 34.5 | 2.27 (m), 1.95 (m) |
| 6 | 27.0 | 1.20–1.85 (overlapped) |
| 7–18 | 29.5–32.5 (overlapped) | 1.20–1.85 (overlapped) |
| 19 | 23.3 | 1.20–1.85 (overlapped) |
| 20 | 14.7 | 0.89 (m) |
| 1′ | 175.6 | |
| 2′ | 72.9 | 4.64 (dd, 10.8, 4.5) |
| 3′ | 36.1 | 2.24 (m), 2.07 (m) |
| 4′ | 26.2 | 1.20–1.85 (overlapped) |
| 5′–20′ | 29.5–32.5 (overlapped) | 1.20–1.85 (overlapped) |
| 21′ | 23.3 | 1.20–1.85 (overlapped) |
| 22′ | 14.7 | 0.89 (m) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Geng, H.; Zhao, C.; Li, F.; Li, Z.-F.; Lun, B.; Wang, C.; Yu, H.; Bie, S.; Li, Z. Chemical Constituents with Inhibitory Activity of NO Production from a Wild Edible Mushroom, Russula vinosa Lindbl, May Be Its Nutritional Ingredients. Molecules 2019, 24, 1305. https://doi.org/10.3390/molecules24071305
Zhang G, Geng H, Zhao C, Li F, Li Z-F, Lun B, Wang C, Yu H, Bie S, Li Z. Chemical Constituents with Inhibitory Activity of NO Production from a Wild Edible Mushroom, Russula vinosa Lindbl, May Be Its Nutritional Ingredients. Molecules. 2019; 24(7):1305. https://doi.org/10.3390/molecules24071305
Chicago/Turabian StyleZhang, Guodong, Huawei Geng, Chunxia Zhao, Fangyi Li, Zhen-Fa Li, Boshu Lun, Chunhua Wang, Heshui Yu, Songtao Bie, and Zheng Li. 2019. "Chemical Constituents with Inhibitory Activity of NO Production from a Wild Edible Mushroom, Russula vinosa Lindbl, May Be Its Nutritional Ingredients" Molecules 24, no. 7: 1305. https://doi.org/10.3390/molecules24071305