Pilot Evaluation of Two Fasciola hepatica Biomarkers for Supporting Triclabendazole (TCBZ) Efficacy Diagnostics
Abstract
:1. Introduction
2. Results
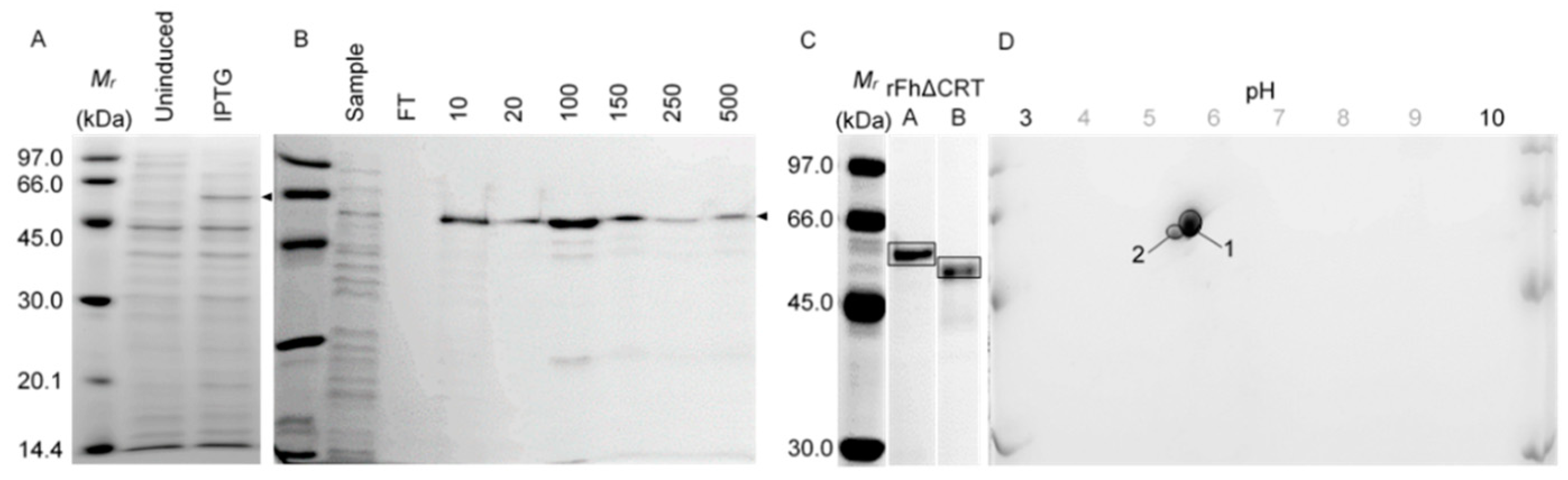
2.1. In Vitro Proteomics: Recombinant Expression and Purification
2.1.1. Calreticulin (N-Terminally Truncated, rFh∆CRT)
2.1.2. Triose Phosphate Isomerase (rFhTPI)
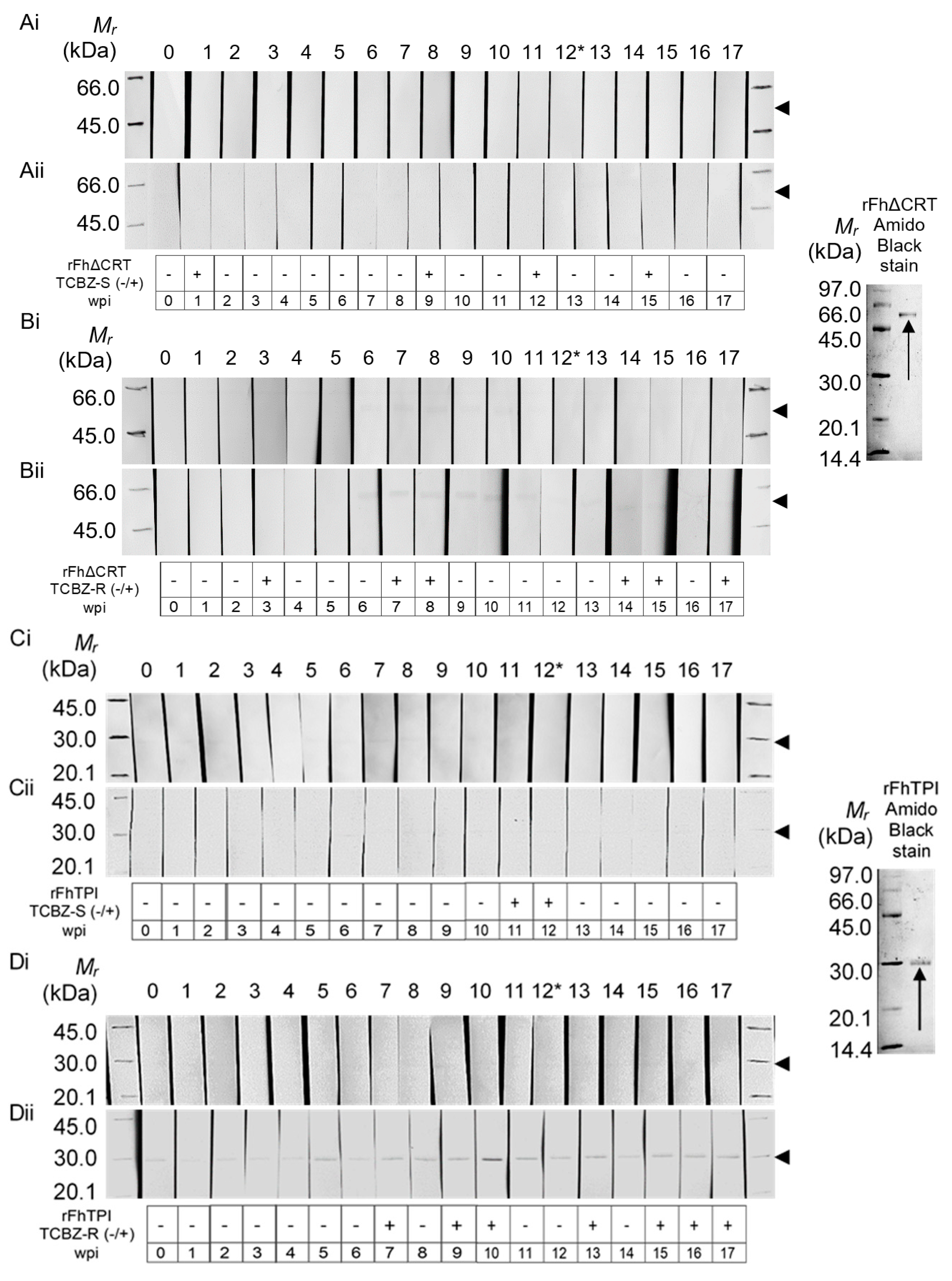
2.2. Western Hybridisation
3. Discussion
4. Materials and Methods
4.1. Cloning and Recombinant Protein Expression
4.2. Proteomics and Western Hybridisation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


References
- Food and Agriculture Organization. Diseases of Domestic Animals Caused by Liver Flukes: Epidemiology, Diagnosis and Control of Fasciola, Paramphistome, Dicroceoelium, Eurytrema and Schistosome Infections of Ruminants in Developing Countries; Food and Agriculture Organization: Rome, Italy, 1994. [Google Scholar]
- Behm, C.A.; Sangster, N.C. Pathology, pathophysiology and clinical aspects. In Fasciolosis; CABI Publishing: Wallingford, UK, 1999; pp. 185–224. [Google Scholar]
- Mas-Coma, S.; Valero, M.A.; Bargues, M.D. Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet. Parasitol. 2009, 163, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Fox, N.J.; White, P.C.L.; Mcclean, C.J.; Marion, G.; Evans, A.; Michael, R. Predicting Impacts of Climate Change on Fasciola hepatica Risk. PLoS ONE 2011, 6, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Boray, J.C.; Crowfoot, P.D.; Strong, M.B.; Allison, J.R.; Schellenbaum, M.; von Orelli, M.; Sarasin, G. Treatment of immature and mature Fasciola hepatica infections in sheep with triclabendazole. Vet. Rec. 1983, 113, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.M.; Elliott, T.P.; Beddoe, T.; Anderson, G.R.; Skuce, P.; Spithill, T.W. Current Threat of Triclabendazole Resistance in Fasciola hepatica. Trends Parasitol. 2016, 32, 458–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamaludeen, J.; Graham-Brown, J.; Stephens, N.; Miller, J.; Howell, A.; Beesley, N.J.; Hodgkinson, J.; Learmount, J.; Williams, D. Lack of efficacy of triclabendazole against Fasciola hepatica is present on sheep farms in three regions of England, and Wales. Vet. Rec. 2019, 184, 502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Hernández, V.; Mulcahy, G.; Pérez, J.; Martínez-Moreno, Á.; Donnelly, S.; Neill, S.M.O.; Dalton, J.P.; Cwiklinski, K. Fasciola hepatica vaccine: We may not be there yet but we’re on the right road. Vet. Parasitol. 2015, 208, 101–111. [Google Scholar] [CrossRef] [Green Version]
- Jefferies, J.R.; Campbell, A.M.; van Rossum, A.J.; Barrett, J.; Brophy, P.M. Proteomic analysis of Fasciola hepatica excretory-Secretory products. Proteomics 2001, 1, 1128–1132. [Google Scholar] [CrossRef]
- Morphew, R.M.; Eccleston, N.; Wilkinson, T.J.; McGarry, J.; Perally, S.; Prescott, M.; Ward, D.; Williams, D.; Paterson, S.; Raman, M.; et al. Proteomics and in silico approaches to extend understanding of the glutathione transferase superfamily of the tropical liver fluke fasciola gigantica. J. Proteome Res. 2012, 11, 5876–5889. [Google Scholar] [CrossRef]
- Morphew, R.M.; Wilkinson, T.J.; Mackintosh, N.; Jahndel, V.; Paterson, S.; McVeigh, P.; Abbas Abidi, S.M.; Saifullah, K.; Raman, M.; Ravikumar, G. Exploring and Expanding the Fatty-Acid-Binding Protein Superfamily in Fasciola Species. J. Proteome Res. 2016, 15, 3308–3321. [Google Scholar] [CrossRef] [Green Version]
- Aguillón, J.C.; Ferreira, L.; Pérez, C.; Colombo, A.; Molina, M.C.; Wallace, A.; Solari, A.; Carvallo, P.; Galindo, M.; Galanti, N.; et al. Tc45, a dimorphic Trypanosoma cruzi immunogen with variable chromosomal localization, is calreticulin. Am. J. Trop. Med. Hyg. 2000, 63, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Groenendyk, J.; Papp, S.; Dabrowska, M.; Knoblach, B.; Kay, C.; Parker, J.M.R.; Opas, M.; Michalak, M. Identification of an N-domain Histidine Essential for Chaperone Function in Calreticulin. J. Biol. Chem. 2003, 278, 50645–50653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, V.; Groenendyk, J.; Steiner, S.S.; Guo, L.; Dabrowska, M.; Parker, J.M.R.; Müller-Esterl, W.; Opas, M.; Michalak, M. Identification by mutational analysis of amino acid residues essential in the chaperone function of calreticulin. J. Biol. Chem. 2006, 281, 2338–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, G.; Pocanschi, C.L.; Rosenauer, A.; Bastos-Aristizabal, S.; Gorelik, A.; Williams, D.B.; Gehring, K. Structural basis of carbohydrate recognition by calreticulin. J. Biol. Chem. 2010, 285, 38612–38620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinsser, V.L.; Hoey, E.M.; Trudgett, A.; Timson, D.J. Biochemical characterisation of triose phosphate isomerase from the liver fluke Fasciola hepatica. Biochimie 2013, 95, 2182–2189. [Google Scholar] [CrossRef] [PubMed]
- Borchert, T.V.; Abagyan, R.A.; Kishan, K.V.R.; Zeelen, J.P.; Wierenga, R.K. The crystal structure of an engineered monomeric triosephosphate isomerase, monoTIM: The correct modelling of an eight-Residue loop. Structure 1993, 1, 205–213. [Google Scholar] [CrossRef]
- Norledge, B.V.; Lambeir, A.M.; Abagyan, R.A.; Rottmann, A.; Fernandez, A.M.; Filimonov, M.; Vladimir, V.; Peter, G.; Wierenga, R.K. Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8)of triosephosphate isomerase: Toward a new substrate specificity. Proteins Struct. Funct. Genet. 2001, 42, 383–389. [Google Scholar] [CrossRef]
- Schliebs, W.; Thanki, N.; Eritja, R.; Wierenga, R. Active site properties of monomeric triosephosphate isomerase (monoTIM) as deduced from mutational and structural studies. Protein Sci. 1996, 5, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Schliebs, W.; Thanki, N.; Jaenicke, R.; Wierenga, R.K. A double mutation at the tip of the dimer interface loop of triosephosphate isomerase generates active monomers with reduced stability. Biochemistry 1997, 36, 9655–9662. [Google Scholar] [CrossRef]
- Walker, S.M.; Johnston, C.; Hoey, E.M.; Fairweather, I.; Borgsteede, F.; Gaasenbeek, C.; Prodöhl, P.A.; Trudgett, A. Population dynamics of the liver fluke, Fasciola hepatica: The effect of time and spatial separation on the genetic diversity of fluke populations in the Netherlands. Parasitology 2011, 138, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Charlier, J.; Ghebretinsae, A.H.; Levecke, B.; Ducheyne, E.; Claerebout, E.; Vercruysse, J. Climate-Driven longitudinal trends in pasture-borne helminth infections of dairy cattle. Int. J. Parasitol. 2016, 46, 881–888. [Google Scholar] [CrossRef]
- Ndao, M. Diagnosis of Parasitic Diseases: Old and New Approaches. Interdiscip. Perspect. Infect. Dis. 2009, 2009, 278246. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sernández, V.; Muiño, L.; Perteguer, M.J.; Gárate, T.; Mezo, M.; González-Warleta, M.; Muro, A.; Correia, d.C.J.M.; Romarís, F.; Ubeira, F.M. Development and evaluation of a new lateral flow immunoassay for serodiagnosis of human fasciolosis. PLoS Negl. Trop. Dis. 2011, 5, e1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, T.R.; Ainsworth, S.; Armstong, S.; Walker, A.; Hodgkinson, S.H.J.; Williams, D. Development of A Pen-Side Diagnostic Test for Liver Fluke Infections in Cattle and Sheep. In British Cattle Conference; University of Liverpool: Liverpool, UK, 2018. [Google Scholar]
- Morphew, R.M.; Mackintosh, N.; Hart, E.H.; Prescott, M.C.; Lacourse, E.J.; Brophy, P.M. In vitro biomarker discovery in the parasitic flatworm Fasciola hepatica for monitoring chemotherapeutic treatment. EuPA Open Proteom. 2014, 3, 85–99. [Google Scholar] [CrossRef]
- González, E.; del Carmen, G.d.L.M.; Meza, I.; Ocadiz-Delgado, R.; Gariglio, P.; Silva-Olivares, A.; Galindo-Gómez, S.; Shibayama, M.; Morán, P.; Valadez, A.; et al. Entamoeba histolytica calreticulin: An endoplasmic reticulum protein expressed by trophozoites into experimentally induced amoebic liver abscesses. Parasitol. Res. 2011, 108, 439–449. [Google Scholar] [CrossRef]
- Ribeiro, C.H.; López, N.C.; Ramírez, G.A.; Valck, C.E.; Molina, M.C.; Aguilar, L.; Rodríguez, M.; Maldonado, I.; Martínez, R.; González, C.; et al. Trypanosoma cruzi calreticulin: A possible role in Chagas’ disease autoimmunity. Mol. Immunol. 2009, 46, 1092–1099. [Google Scholar] [CrossRef]
- Ferreira, V.; Molina, M.C.; Valck, C.; Rojas, Á.; Aguilar, L.; Ramírez, G.; Schwaeble, W.; Ferreira, A. Role of calreticulin from parasites in its interaction with vertebrate hosts. Mol. Immunol. 2004, 40, 1279–1291. [Google Scholar] [CrossRef]
- Chou, H.C.; Chen, J.Y.; Lin, D.Y.; Wen, Y.F.; Lin, C.C.; Lin, S.H.; Lin, C.H.; Chung, T.W.; Liao, E.C.; Chen, Y.J.; et al. Identification of up- and down-Regulated proteins in pemetrexed-Resistant human lung adenocarcinoma: Flavin reductase and calreticulin play key roles in the development of pemetrexed-associated resistance. J. Proteome Res. 2015, 14, 4907–4920. [Google Scholar] [CrossRef]
- Kopecka, J.; Campia, I.; Brusa, D.; Doublier, S.; Matera, L.; Ghigo, D.; Bosia, A.; Riganti, C. Nitric oxide and P-Glycoprotein modulate the phagocytosis of colon cancer cells. J. Cell. Mol. Med. 2011, 15, 1492–1504. [Google Scholar] [CrossRef]
- Matsukuma, S.; Yoshimura, K.; Ueno, T.; Oga, A.; Inoue, M.; Watanabe, Y.; Kuramasu, A.; Fuse, M.; Tsunedomi, R.; Nagaoka, S.; et al. Calreticulin is highly expressed in pancreatic cancer stem-Like cells. Cancer Sci. 2016, 107, 1599–1609. [Google Scholar] [CrossRef]
- Hernández-González, A.; Valero, M.L.L.; Pino, M.S.d.; Oleaga, A.; Siles-Lucas, M. Proteomic analysis of in vitro newly excysted juveniles from Fasciola hepatica. Mol. Biochem. Parasitol. 2010, 172, 121–128. [Google Scholar] [CrossRef]
- Chemale, G.; Perally, S.; Lacourse, E.J.; Prescott, M.C.; Jones, L.M.; Ward, D.; Meaney, M.; Hoey, E.; Brennan, G.P.; Fairweather, I.; et al. Comparative proteomic analysis of triclabendazole response in the liver fluke fasciola hepatica. J. Proteome Res. 2010, 9, 4940–4951. [Google Scholar] [CrossRef]
- Gelebart, P.; Opas, M.; Michalak, M. Calreticulin, a Ca2+-binding chaperone of the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 2005, 37, 260–266. [Google Scholar] [CrossRef]
- Michalak, M.; Mariani, P.; Opas, M. Calreticulin, a multifunctional Ca2+ binding chaperone of the endoplasmic reticulum. Biochem. Cell Biol. Biol. Cell. 1998, 76, 779–785. [Google Scholar] [CrossRef]
- Park, B.J.; Lee, D.G.; Yu, J.R.; Jung, S.K.; Choi, K.; Lee, J.; Lee, J.; Kim, Y.S.; Lee, J.I.; Kwon, J.Y.; et al. Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol. Biol. Cell 2001, 12, 2835–2845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedard, K.; Szabo, E.; Michalak, M.; Opas, M. Cellular functions of endoplasmic reticulum chaperones calreticulin, calnexin, and ERp57. Int. Rev. Cytol. 2005, 245, 91–121. [Google Scholar] [PubMed]
- Cribb, A.E.; Peyrou, M.; Muruganandan, S.; Schneider, L. The endoplasmic reticulum in xenobiotic toxicity. Drug Metab. Rev. 2005, 37, 405–442. [Google Scholar] [CrossRef] [PubMed]
- Bedard, K.; MacDonald, N.; Collins, J.; Cribb, A. Cytoprotection Following Endoplasmic Reticulum Stress Protein Induction in Continuous Cell Lines. Basic Clin. Pharmacol. Toxicol. 2004, 94, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, D.; de la Rubia, J.E.; Carrasco-Abad, A.M.; Toledo, R.; Mas-Coma, S.; Marcilla, A. Identification of enolase as a plasminogen-Binding protein in excretory-Secretory products of Fasciola hepatica. FEBS Lett. 2004, 563, 203–206. [Google Scholar] [CrossRef] [Green Version]
- Pancholi, V.; Chhatwal, G.S. Housekeeping enzymes as virulence factors for pathogens. Int. J. Med. Microbiol. 2003, 293, 391–401. [Google Scholar] [CrossRef]
- Seigle, J.L.; Celotto, A.M.; Palladino, M.J. Degradation of functional triose phosphate isomerase protein underlies sugarkill pathology. Genetics 2008, 179, 855–862. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Davis, C.N.; Phillips, H.; Tomes, J.J.; Swain, M.T.; Wilkinson, T.J.; Brophy, P.M.; Morphew, R.M. The importance of extracellular vesicle purification for downstream analysis: A comparison of differential centrifugation and size exclusion chromatography for helminth pathogens. PLoS Negl. Trop. Dis. 2019, 13, e0007191. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-Friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Varghese, A.; Solanki, J.B. Seroprevalence of Fasciola gigantica infection in bovines using cysteine proteinase dot enzyme-Linked immunosorbent assay. Vet. World 2017, 10, 1189–1193. [Google Scholar] [CrossRef] [Green Version]
- Raadsma, H.W.; Kingsford, N.M.; Suharyanta; Spithill, T.W.; Piedrafita, D. Host responses during experimental infection with Fasciola gigantica or Fasciola hepatica in Merino sheep. I. Comparative immunological and plasma biochemical changes during early infection. Vet. Parasitol. 2007, 143, 275–286. [Google Scholar] [CrossRef]
- Michalak, M.; Corbett, E.F.; Mesaeli, N.; Nakamura, K.; Opas, M. Calreticulin: One protein, one gene, many functions. Biochem. J. 1999, 344, 281–292. [Google Scholar] [CrossRef]
Sample Availability: Samples of primers, plasmid, or recombinant cell-lines are available from R.M.M. and P.M.B. authors upon request. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Collett, C.F.; Morphew, R.M.; Timson, D.; Phillips, H.C.; Brophy, P.M. Pilot Evaluation of Two Fasciola hepatica Biomarkers for Supporting Triclabendazole (TCBZ) Efficacy Diagnostics. Molecules 2020, 25, 3477. https://doi.org/10.3390/molecules25153477
Collett CF, Morphew RM, Timson D, Phillips HC, Brophy PM. Pilot Evaluation of Two Fasciola hepatica Biomarkers for Supporting Triclabendazole (TCBZ) Efficacy Diagnostics. Molecules. 2020; 25(15):3477. https://doi.org/10.3390/molecules25153477
Chicago/Turabian StyleCollett, Clare F., Russell M. Morphew, David Timson, Helen C. Phillips, and Peter M. Brophy. 2020. "Pilot Evaluation of Two Fasciola hepatica Biomarkers for Supporting Triclabendazole (TCBZ) Efficacy Diagnostics" Molecules 25, no. 15: 3477. https://doi.org/10.3390/molecules25153477







