Application of QCM in Peptide and Protein-Based Drug Product Development
Abstract
:1. Introduction
2. QCM Technology Basics
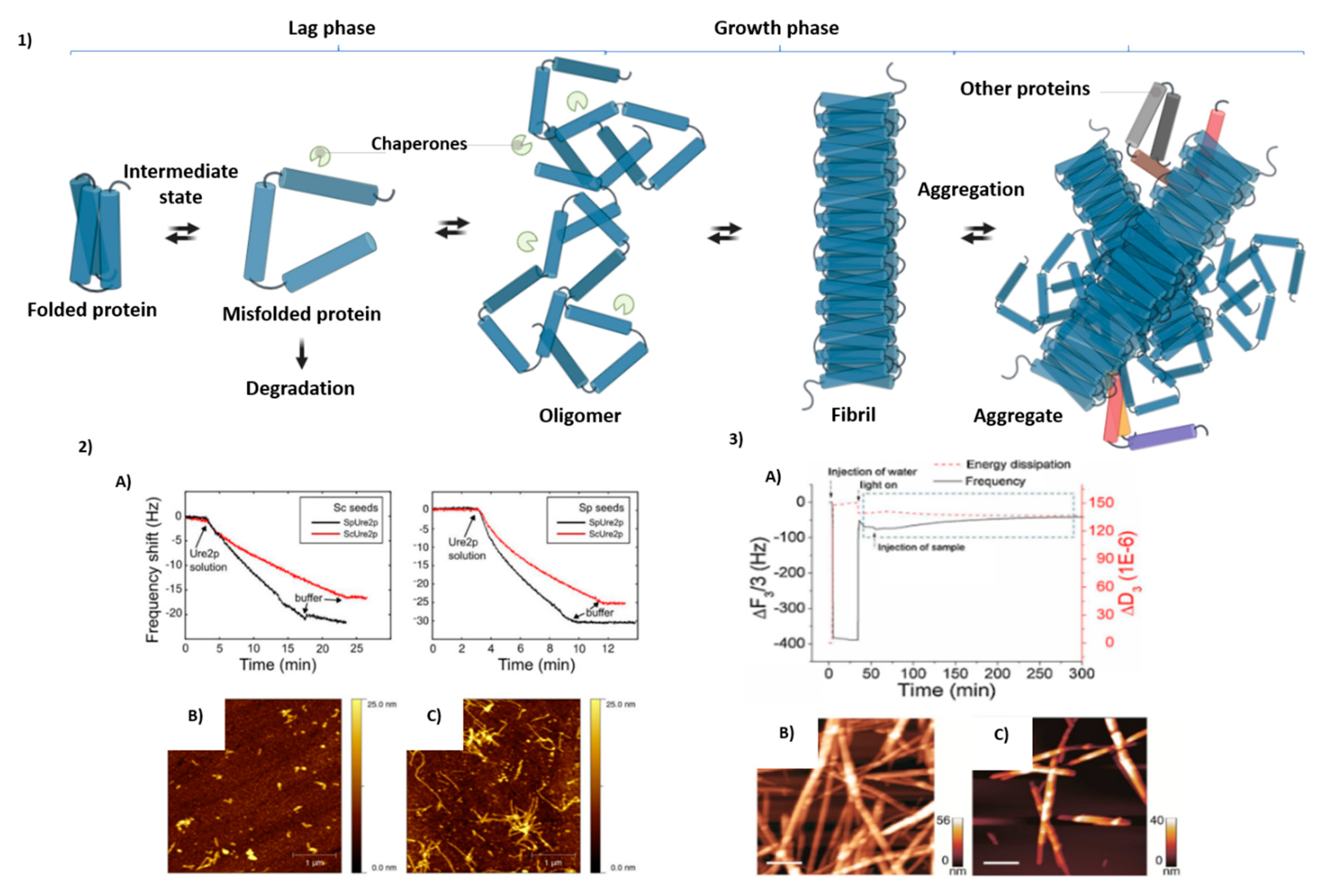
3. Adsorption of Peptide and Protein Molecules on the QCM Surface
4. Application of the QCM in Formulation and Primary Packaging Development
5. Application of the QCM in Drug Product Manufacturing Process Development
6. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Moorkens, E.; Meuwissen, N.; Huys, I.; Declerck, P.; Vulto, A.G.; Simoens, S. The market of biopharmaceutical medicines: A snapshot of a diverse industrial landscape. Front. Pharmacol. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urquhart, L. Top Companies and Drugs by Sales in 2019. Available online: https://www.nature.com/articles/d41573-020-00047-7 (accessed on 28 June 2020).
- Moorkens, E.; Jonker-Exler, C.; Huys, I.; Declerck, P.; Simoens, S.; Vulto, A.G. Overcoming barriers to the market access of biosimilars in the European Union: The case of biosimilar monoclonal antibodies. Front. Pharmacol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weert, V. Pharmaceutical Formulation Development of Peptides and Proteins; CRC Press: Boca Raton, FL, USA, 1999; ISBN 9781439853894. [Google Scholar]
- Nema, S.; Ludwig, J.D. Parenteral Medications; CRC Press: Boca Raton, FL, USA, 2019; ISBN 9788578110796. [Google Scholar]
- Shire, S.J. Monoclonal Antibodies Meeting the Challenges in Manufacturing, Formulation, Delivery and Stability of Final Drug Product; Elsevier: Cambridge, UK, 2015; ISBN 9788578110796. [Google Scholar]
- Shah, M. Commentary: New perspectives on protein aggregation during Biopharmaceutical development. Int. J. Pharm. 2018, 552, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mahler, H.C.; Jiskoot, W. Analysis of Aggregates and Particles in Protein Pharmaceuticals; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; ISBN 9780470497180. [Google Scholar]
- Zurdo, J.; Michael, R.; Stallwood, Y.; Hedman, K.; Aastrup, T. Improving the developability of biopharmaceuticals. Innov. Pharm. Technol. 2011, 37, 34–39. [Google Scholar]
- Sunar, M. 2.22 Piezoelectric materials. In Comprehensive Energy Systems; Elsevier: Cambridge, MA, USA, 2018; ISBN 9780128095973. [Google Scholar]
- Reyes, P.I.; Duan, Z.; Lu, Y.; Khavulya, D.; Boustany, N. ZnO nanostructure-modified QCM for dynamic monitoring of cell adhesion and proliferation. Biosens. Bioelectron. 2013, 41, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, T.; Szulczyński, B.; Kamysz, W.; Gębicki, J.; Namieśnik, J. Evaluation of three peptide immobilization techniques on a QCM surface related to acetaldehyde responses in the gas phase. Sensors 2018, 18, 3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Huang, Y.; Jin, X.; Mason, A.J.; Zeng, X. Ionic liquid thin layer EQCM explosives sensor. Sens. Actuators B Chem. 2009, 140, 363–370. [Google Scholar] [CrossRef]
- Jha, S.K.; Hayashi, K. A quick responding quartz crystal microbalance sensor array based on molecular imprinted polyacrylic acids coating for selective identification of aldehydes in body odor. Talanta 2015, 134, 105–119. [Google Scholar] [CrossRef]
- Huang, X.-H.; Bai, Q.; Hu, J.; Hou, D. A practical model of quartz crystal microbalance in actual applications. Sensors 2017, 17, 1785. [Google Scholar] [CrossRef] [Green Version]
- Avila, J.R.; Demarco, E.J.; Emery, J.D.; Farha, O.K.; Pellin, M.; Hupp, J.T.; Martinson, A.B.F. Real-time observation of atomic layer deposition inhibition: Metal oxide growth on self-assembled alkanethiols. ACS Appl. Mater. Interfaces 2014, 6, 11891–11898. [Google Scholar] [CrossRef]
- Latif, U.; Can, S.; Hayden, O.; Grillberger, P.; Dickert, F.L. Sauerbrey and anti-Sauerbrey behavioral studies in QCM sensors—Detection of bioanalytes. Sens. Actuators B Chem. 2013, 176, 825–830. [Google Scholar] [CrossRef]
- Muckley, E.S.; Anazagasty, C.; Jacobs, C.B.; Hianik, T.; Ivanov, I.N. Low-cost scalable quartz crystal microbalance array for environmental sensing. In Proceedings of the Organic Sensors and Bioelectronics IX; SPIE-International Society Optical Engineering: San Diego, CA, USA, 2016; Volume 9944, p. 99440Y. [Google Scholar]
- Beißner, S.; Thies, J.-W.; Bechthold, C.; Kuhn, P.; Thürmann, B.; Dübel, S.; Dietzel, A. Low-cost, in-liquid measuring system using a novel compact oscillation circuit and quartz-crystal microbalances (QCMs) as a versatile biosensor platform. J. Sens. Sens. Syst. 2017, 6, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Tao, X.; Feng, B.; Yu, L.; Wang, D.; Dong, S.; Luo, J. A humidity sensor based on quartz crystal microbalance using graphene oxide as a sensitive layer. Vacuum 2017, 140, 101–105. [Google Scholar] [CrossRef]
- Yao, C.; Qu, L.; Fu, W.-L. Detection of fibrinogen and coagulation factor VIII in plasma by a quartz crystal microbalance biosensor. Sensors 2013, 13, 6946–6956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Song, S.; Shuai, Q.; Pei, Y.; Aastrup, T.; Pei, Y.; Pei, Z. Real-time and label-free analysis of binding thermodynamics of carbohydrate-protein interactions on unfixed cancer cell surfaces using a QCM biosensor. Sci. Rep. 2015, 5, 14066. [Google Scholar] [CrossRef]
- Zainuddin, A.A.; Nordin, A.N.; Mansor, A.F.M.; Ab Rahim, R.; Mak, W.C. Integrated multichannel electrochemical–quartz crystal microbalance sensors for liquid sensing. IEEE Access 2020, 8, 3668–3676. [Google Scholar] [CrossRef]
- Ogi, H.; Fukukshima, M.; Hamada, H.; Noi, K.; Hirao, M.; Yagi, H.; Goto, Y. Ultrafast propagation of β-amyloid fibrils in oligomeric cloud. Sci. Rep. 2014, 4, 6960. [Google Scholar] [CrossRef]
- Hajiraissi, R.; Hanke, M.; Yang, Y.; Duderija, B.; Orive, A.G.; Grundmeier, G.; Keller, A. Adsorption and fibrillization of islet amyloid polypeptide at self-assembled monolayers studied by QCM-D, AFM, and PM-IRRAS. Langmuir 2018, 34, 3517–3524. [Google Scholar] [CrossRef]
- Wasilewski, T.; Szulczyński, B.; Wojciechowski, M.; Kamysz, W.; Gębicki, J. Determination of long-chain aldehydes using a novel quartz crystal microbalance sensor based on a biomimetic peptide. Microchem. J. 2020, 154, 104509. [Google Scholar] [CrossRef]
- Roto, R.; Rianjanu, A.; Rahmawati, A.; Fatyadi, I.A.; Yulianto, N.; Majid, N.; Syamsu, I.; Wasisto, H.S.; Triyana, K. Quartz crystal microbalances functionalized with citric acid-doped polyvinyl acetate nanofibers for ammonia sensing. ACS Appl. Nano Mater. 2020, 3, 5687–5697. [Google Scholar] [CrossRef]
- Pauliukaite, R.; Voitechovic, E. Multisensor systems and arrays for medical applications employing naturally-occurring compounds and materials. Sensors 2020, 20, 3551. [Google Scholar] [CrossRef] [PubMed]
- Santos-Martinez, M.J.; Inkielewicz-Stępniak, I.; Medina, C.; Rahme, K.; D’Arcy, D.M.; Fox, D.; Holmes, J.D.; Zhang, H.; Radomski, M.W. The use of quartz crystal microbalance with dissipation (QCM-D) for studying nanoparticle-induced platelet aggregation. Int. J. Nanomed. 2012, 7, 243–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, S.S.; Chan, H.; Sorci, M.; Van Deventer, J.; Wittrup, K.D.; Belfort, G.; Walt, D.R. Detection of amyloid β oligomers toward early diagnosis of Alzheimer’s disease. Anal. Biochem. 2019, 566, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Ansorena, P.; Zuzuarregui, A.; Pérez-Lorenzo, E.; Mujika, M.; Arana, S. Comparative analysis of QCM and SPR techniques for the optimization of immobilization sequences. Sens. Actuators B Chem. 2011, 155, 667–672. [Google Scholar] [CrossRef]
- Ahumada, L.A.C.; Pérez, N.P.; Sandoval, O.H.; Guerrero, F.D.P.; Olmedo, J.J.S. A new way to find dielectric properties of liquid sample using the quartz crystal resonator (QCR). Sens. Actuators A Phys. 2016, 239, 153–160. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Devlin, G.L.; Dobson, C.M.; Welland, M.E. Probing Protein Aggregation with Quartz Crystal Microbalances. In Protein Folding, Misfolding, and Disease; Hill, A.F., Barnham, K.J., Bottomley, S.P., Cappai, R., Eds.; Humana Press: Totowa, NJ, USA, 2011; Volume 752, ISBN 978-1-60327-221-6. [Google Scholar]
- Gao, J.; Huang, X.-H.; Wang, Y. The modified design of ring electrode quartz crystal resonator for uniform mass sensitivity distribution. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2013, 60, 2031–2034. [Google Scholar] [CrossRef]
- Alassi, A.; Benammar, M.; Brett, D.J.L. Quartz crystal microbalance electronic interfacing systems: A review. Sensors 2017, 17, 2799. [Google Scholar] [CrossRef] [Green Version]
- Bragazzi, N.L.; Amicizia, D.; Panatto, D.; Tramalloni, D.; Valle, I.; Gasparini, R. Quartz-Crystal Microbalance (QCM) for public health. In Advances in Protein Chemistry and Structural Biology; Elsevier: San Diego, CA, USA, 2015; Volume 101, pp. 149–211. [Google Scholar]
- Arif, S.; Qudsia, S.; Urooj, S.; Chaudry, N.; Arshad, A.; Andleeb, S. Blueprint of quartz crystal microbalance biosensor for early detection of breast cancer through salivary autoantibodies against ATP6AP1. Biosens. Bioelectron. 2015, 65, 62–70. [Google Scholar] [CrossRef]
- Atay, S.; Çakır, C.; Yavuz, H.; Pişkin, K.; Yılmaz, F.; Denizli, A. Quartz crystal microbalance based biosensors for detecting highly metastatic breast cancer cells via their transferrin receptors. Anal. Methods 2016, 8, 153–161. [Google Scholar] [CrossRef]
- Sinn, M.H.S.; Hussain, M.; Zeilinger, M.; Northoff, H.; Lieberzeit, P.A.; Gehring, F.K. Blood coagulation thromboplastine time measurements on a nanoparticle coated quartz crystal microbalance biosensor in excellent agreement with standard clinical methods. J. Biosens. Bioelectron. 2013, 4, 1–6. [Google Scholar] [CrossRef]
- Afzal, A.; Mujahid, A.; Schirhagl, R.; Bajwa, S.Z.; Latif, U.; Feroz, S. Gravimetric viral diagnostics: QCM based biosensors for early detection of viruses. Chemosensors 2017, 5, 7. [Google Scholar] [CrossRef]
- Wang, L.; Wang, R.; Chen, F.; Jiang, T.; Wang, H.; Slavik, M.; Wei, H.; Li, Y. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017, 221, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Chen, J.Y. Quartz crystal microbalance in cell biology studies. J. Biochips Tissue Chips 2013, 2013, 5. [Google Scholar] [CrossRef]
- Henstridge, C.M.; Hyman, B.T.; Spires-Jones, T.L. Beyond the neuron–cellular interactions early in Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 2019, 20, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Zeng, X. Monitoring the cellular binding events with Quartz Crystal Microbalance (QCM) biosensors. Adv. Struct. Saf. Stud. 2017, 1572, 313–326. [Google Scholar] [CrossRef]
- Tonda-Turo, C.; Carmagnola, I.; Ciardelli, G. Quartz crystal microbalance with dissipation monitoring: A powerful method to predict the in vivo behavior of bioengineered surfaces. Front. Bioeng. Biotechnol. 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Quesada, A.; Schofield, M.M.; Tsortos, A.; Mateos-Gil, P.; Milioni, D.; Gizeli, E.; Delgado-Buscalioni, R. Hydrodynamics of quartz-crystal-microbalance DNA sensors based on liposome amplifiers. Phys. Rev. Appl. 2020, 13, 064059. [Google Scholar] [CrossRef]
- Pirich, C.L.; De Freitas, R.A.; Torresi, R.M.; Picheth, G.F.; Sierakowski, M.R. Piezoelectric immunochip coated with thin films of bacterial cellulose nanocrystals for dengue detection. Biosens. Bioelectron. 2017, 92, 47–53. [Google Scholar] [CrossRef]
- Al-Husseini, J.K.; Stanton, N.J.; Selassie, C.R.; Johal, M.S. The binding of drug molecules to serum albumin: The effect of drug hydrophobicity on binding strength and protein desolvation. Langmuir 2019, 35, 17054–17060. [Google Scholar] [CrossRef]
- Tagaya, M. In situ QCM-D study of nano-bio interfaces with enhanced biocompatibility. Polym. J. 2015, 47, 599–608. [Google Scholar] [CrossRef]
- Campos, J.; Jimenez, C.; Trigo, C.; Ibarra, P.; Rana, D.; Thiruganesh, R.; Ramalingam, M.; Haidar, Z.S. Quartz crystal microbalance with dissipation monitoring: A powerful tool for bionanoscience and drug discovery. J. Bionanosci. 2015, 9, 249–260. [Google Scholar] [CrossRef]
- Clegg, J.R.; Ludolph, C.M.; Peppas, N.A. QCM-D assay for quantifying the swelling, biodegradation, and protein adsorption of intelligent nanogels. J. Appl. Polym. Sci. 2020, 137, 48655. [Google Scholar] [CrossRef]
- Hampitak, P.; Melendrez, D.; Iliut, M.; Fresquet, M.; Parsons, N.; Spencer, B.; A Jowitt, T.; Vijayaraghavan, A. Protein interactions and conformations on graphene-based materials mapped using a quartz-crystal microbalance with dissipation monitoring (QCM-D). Carbon 2020, 165, 317–327. [Google Scholar] [CrossRef]
- Kolev, I.N.; Ivanova, N.A.; Marinov, M.K.; Alexieva, G.; Strashilov, V. A QCM-based assay of drug content in Eudragit RS 100-based delivery systems. Talanta 2019, 202, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Penn, L.S.; Xi, J. Quartz crystal microbalance: Sensing cell-substrate adhesion and beyond. Biosens. Bioelectron. 2018, 99, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Neupane, S.; De Smet, Y.; Renner, F.U.; Losada-Pérez, P. Quartz crystal microbalance with dissipation monitoring: A versatile tool to monitor phase transitions in biomimetic membranes. Front. Mater. 2018, 5, 46. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, A.J.M.; Oliveira, A.R.; Roque, A.C.A. Protein- and peptide-based biosensors in artificial olfaction. Trends Biotechnol. 2018, 36, 1244–1258. [Google Scholar] [CrossRef] [Green Version]
- Cho, N.-J.; Frank, C.W.; Kasemo, B.; Höök, F. Quartz crystal microbalance with dissipation monitoring of supported lipid bilayers on various substrates. Nat. Protoc. 2010, 5, 1096–1106. [Google Scholar] [CrossRef]
- Baltus, R.E.; Carmon, K.S.; Luck, L.A. Quartz Crystal Microbalance (QCM) with immobilized protein receptors: Comparison of response to ligand binding for direct protein immobilization and protein attachment via disulfide linker. Langmuir 2007, 23, 3880–3885. [Google Scholar] [CrossRef]
- Ozboyaci, M.; Kokh, D.B.; Corni, S.; Wade, R.C. Modeling and simulation of protein–surface interactions: Achievements and challenges. Q. Rev. Biophys. 2016, 49, 1–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Wang, X.; Wang, L.; Hou, X.; Liu, W.; Chen, C. Interaction of gold nanoparticles with proteins and cells. Sci. Technol. Adv. Mater. 2015, 16, 34610. [Google Scholar] [CrossRef] [PubMed]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein binding pocket dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marx, K.A. Quartz crystal microbalance: A useful tool for studying thin polymer films and complex biomolecular systems at the solution−surface interface. Biomacromolecules 2003, 4, 1099–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapadka, K.L.; Becher, F.J.; Dos Santos, A.L.G.; Jackson, S.E. Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus 2017, 7, 20170030. [Google Scholar] [CrossRef] [Green Version]
- Roberts, C.J. Protein aggregation and its impact on product quality. Curr. Opin. Biotechnol. 2014, 30, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Boil. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Chuang, E.; Hori, A.; Hesketh, C.D.; Shorter, J. Amyloid assembly and disassembly. J. Cell Sci. 2018, 131, jcs189928. [Google Scholar] [CrossRef] [Green Version]
- Marshall, K.E.; Marchante, R.; Xue, W.-F.; Serpell, L.C. The relationship between amyloid structure and cytotoxicity. Prion 2014, 8, 192–196. [Google Scholar] [CrossRef] [Green Version]
- Deger, J.M.; Gerson, J.; Kayed, R. The interrelationship of proteasome impairment and oligomeric intermediates in neurodegeneration. Aging Cell 2015, 14, 715–724. [Google Scholar] [CrossRef]
- Fusco, G.; Chen, S.; Williamson, P.T.; Cascella, R.; Perni, M.; Jarvis, J.A.; Cecchi, C.; Vendruscolo, M.; Chiti, F.; Cremades, N.; et al. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science 2017, 358, 1440–1443. [Google Scholar] [CrossRef] [Green Version]
- Langkilde, A.; Vestergaard, B. Methods for structural characterization of prefibrillar intermediates and amyloid fibrils. FEBS Lett. 2009, 583, 2600–2609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, S.; Barnett, G.V.; Pathak, J.A.; Roberts, C.J.; Sarangapani, P.S. Protein aggregation, particle formation, characterization & rheology. Curr. Opin. Colloid Interface Sci. 2014, 19, 438–449. [Google Scholar] [CrossRef] [Green Version]
- Herczenik, E.; Gebbink, M.F.B.G. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008, 22, 2115–2133. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.W.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [Green Version]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Boil. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Watt, B.; Van Niel, G.; Raposo, G.; Marks, M.S. PMEL: A pigment cell-specific model for functional amyloid formation. Pigment Cell Melanoma Res. 2013, 26, 300–315. [Google Scholar] [CrossRef] [Green Version]
- Patterson, C. Alzheimers Disease International World Alzheimer’s Report 2018; Alzheimer’s Disease International: London, UK, 2018; ISBN 9783804728387. [Google Scholar]
- Wang, Y.-Q.; Buell, A.K.; Wang, X.-Y.; Welland, M.E.; Dobson, C.M.; Knowles, T.P.J.; Perrett, S. Relationship between prion propensity and the rates of individual molecular steps of fibril assembly. J. Boil. Chem. 2011, 286, 12101–12107. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Feng, Y.; Tian, X.; Li, C.; Liu, L. Disassembling and degradation of amyloid protein aggregates based on gold nanoparticle-modified g-C3N4. Colloids Surf. B Biointerfaces 2020, 192, 111051. [Google Scholar] [CrossRef]
- Kotarek, J.; Moss, M. Impact of phospholipid bilayer saturation on amyloid-β protein aggregation intermediate growth: A quartz crystal microbalance analysis. Anal. Biochem. 2010, 399, 30–38. [Google Scholar] [CrossRef]
- Kotarek, J.A.; Johnson, K.C.; Moss, M.A. Quartz crystal microbalance analysis of growth kinetics for aggregation intermediates of the amyloid-β protein. Anal. Biochem. 2008, 378, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Shu, W.; Devlin, G.L.; Meehan, S.; Auer, S.; Dobson, C.M.; Welland, M.E. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc. Natl. Acad. Sci. USA 2007, 104, 10016–10021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovgaard, M.B.; Dong, M.; Otzen, D.E.; Besenbacher, F. Quartz crystal microbalance studies of multilayer glucagon fibrillation at the solid-liquid interface. Biophys. J. 2007, 93, 2162–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, M.; Nabok, A.; Parkinson, D.; Tothill, I.; Salam, F.; Tsargorodskaya, A. Detection of β-amyloid peptide (1–16) and amyloid precursor protein (APP770) using spectroscopic ellipsometry and QCM techniques: A step forward towards Alzheimers disease diagnostics. Biosens. Bioelectron. 2010, 26, 1332–1336. [Google Scholar] [CrossRef] [Green Version]
- Ragaliauskas, T.; Mickevicius, M.; Budvytyte, R.; Niaura, G.; Carbonnier, B.; Valincius, G. Adsorption of β-amyloid oligomers on octadecanethiol monolayers. J. Colloid Interface Sci. 2014, 425, 159–167. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Wang, J.; Feng, Y.; Xu, E.; Mao, X.; Liu, L. Evaluation of the photo-degradation of Alzheimer’s amyloid fibrils with a label-free approach. Chem. Commun. 2018, 54, 13084–13087. [Google Scholar] [CrossRef]
- Gaspar, R.; Meisl, G.; Young, L.; Kaminski, C.F.; Knowles, T.P.J.; Sparr, E.; Linse, S.; Buell, A.K. Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 2017, 50, e6. [Google Scholar] [CrossRef] [Green Version]
- Kametani, F.; Hasegawa, M. Reconsideration of amyloid hypothesis and tau hypothesis in Alzheimer’s disease. Front. Mol. Neurosci. 2018, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Ke, P.C.; Pilkington, E.H.; Sun, Y.; Javed, I.; Kakinen, A.; Peng, G.; Ding, F.; Davis, T.P. Mitigation of amyloidosis with nanomaterials. Adv. Mater. 2019, 32, e1901690. [Google Scholar] [CrossRef]
- Keller, A.; Grundmeier, G. Amyloid aggregation at solid-liquid interfaces: Perspectives of studies using model surfaces. Appl. Surf. Sci. 2020, 506, 144991. [Google Scholar] [CrossRef]
- Nilsson, M.R. Techniques to study amyloid fibril formation in vitro. Methods 2004, 34, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Zheng, Q. Applications of mass spectrometry in the onset of amyloid fibril formation: Focus on the analysis of early-stage oligomers. Front. Chem. 2020, 8, 324. [Google Scholar] [CrossRef] [PubMed]
- Cole, H.; Porrini, M.; Morris, R.J.; Smith, T.; Kalapothakis, J.; Weidt, S.; Mackay, C.L.; Macphee, C.E.; Barran, P.E. Early stages of insulin fibrillogenesis examined with ion mobility mass spectrometry and molecular modelling. Analyst 2015, 140, 7000–7011. [Google Scholar] [CrossRef] [Green Version]
- Ausili, A.; Berglin, M.; Elwing, H.; Corbalán-García, S.; Gómez-Fernández, J. Quartz crystal microbalance with dissipation monitoring and the real-time study of biological systems and macromolecules at interfaces. Biomed. Spectrosc. Imaging 2012, 1, 325–338. [Google Scholar] [CrossRef]
- Rodriguez, R.A.; Chen, L.; Plascencia-Villa, G.; Perry, G. Thermodynamics of amyloid-β fibril elongation: Atomistic details of the transition state. ACS Chem. Neurosci. 2017, 9, 783–789. [Google Scholar] [CrossRef]
- Buell, A.K.; Dobson, C.M.; Welland, M.E. Measuring the kinetics of amyloid fibril elongation using quartz crystal microbalances. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Singapore, Singapore, 2012; Volume 849, pp. 101–119. [Google Scholar]
- Westwood, M.; Kirby, A.R.; Parker, R.; Morris, V.J. Combined QCMD and AFM studies of lysozyme and poly-l-lysine–poly-galacturonic acid multilayers. Carbohydr. Polym. 2012, 89, 1222–1231. [Google Scholar] [CrossRef]
- Zanden, C.M.V.; Wampler, L.; Bowers, I.; Watkins, E.B.; Majewski, J.; Chi, E.Y. Fibrillar and nonfibrillar amyloid beta structures drive two modes of membrane-mediated toxicity. Langmuir 2019, 35, 16024–16036. [Google Scholar] [CrossRef]
- Hajiraissi, R.; Hanke, M.; Orive, A.G.; Duderija, B.; Hofmann, U.; Zhang, Y.; Grundmeier, G.; Keller, A. Effect of terminal modifications on the adsorption and assembly of hIAPP(20–29). ACS Omega 2019, 4, 2649–2660. [Google Scholar] [CrossRef]
- Kubiak, K.; Adamczyk, Z.; Wasilewska, M. Mechanisms of fibrinogen adsorption at the silica substrate determined by QCM-D measurements. J. Colloid Interface Sci. 2015, 457, 378–387. [Google Scholar] [CrossRef]
- Kabay, G.; Can, G.K.; Mutlu, M. Amyloid-like protein nanofibrous membranes as a sensing layer infrastructure for the design of mass-sensitive biosensors. Biosens. Bioelectron. 2017, 97, 285–291. [Google Scholar] [CrossRef]
- Stroo, E.; Koopman, M.; Nollen, E.A.A.; Mata-Cabana, A. Cellular regulation of amyloid formation in aging and disease. Front. Mol. Neurosci. 2017, 11, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, D.; Mahammad, S.S.; Singh, P.P.; Kodipyaka, R. A review on parenteral delivery of peptides and proteins. Drug Dev. Ind. Pharm. 2019, 45, 1403–1420. [Google Scholar] [CrossRef] [PubMed]
- Moussa, E.M.; Panchal, J.P.; Moorthy, B.S.; Blum, J.S.; Joubert, M.K.; Narhi, L.O.; Topp, E.M. Immunogenicity of therapeutic protein aggregates. J. Pharm. Sci. 2016, 105, 417–430. [Google Scholar] [CrossRef] [Green Version]
- Warne, N.W.; Mahler, H.-C. Challenges in Protein Product Development; Springer International Publishing AG: Cham, Switzerland, 2018; ISBN 9783319906010. [Google Scholar]
- Hartl, E.; Dixit, N.; Besheer, A.; Kalonia, D.; Winter, G. Weak antibody–cyclodextrin interactions determined by quartz crystal microbalance and dynamic/static light scattering. Eur. J. Pharm. Biopharm. 2013, 85, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Funke, S.; Matilainen, J.; Nalenz, H.; Bechtold-Peters, K.; Mahler, H.-C.; Friess, W. Optimization of the bake-on siliconization of cartridges. Part I: Optimization of the spray-on parameters. Eur. J. Pharm. Biopharm. 2016, 104, 200–215. [Google Scholar] [CrossRef]
- Höger, K.; Mathes, J.; Fries, W. IgG1 adsorption to siliconized glass vials—Influence of pH, ionic strength, and nonionic surfactants. J. Pharm. Sci. 2015, 104, 34–43. [Google Scholar] [CrossRef]
- Thirumangalathu, R.; Krishnan, S.; Ricci, M.S.; Brems, D.N.; Randolph, T.W.; Carpenter, J.F. Silicone oil- and agitation-induced aggregation of a monoclonal antibody in aqueous solution. J. Pharm. Sci. 2009, 98, 3167–3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, L.S.; Kaufmann, A.; Middaugh, C.R. Silicone oil induced aggregation of proteins. J. Pharm. Sci. 2005, 94, 918–927. [Google Scholar] [CrossRef]
- Dixit, N.; Maloney, K.M.; Kalonia, D.S. Application of quartz crystal microbalance to study the impact of pH and ionic strength on protein–silicone oil interactions. Int. J. Pharm. 2011, 412, 20–27. [Google Scholar] [CrossRef]
- Dixit, N.; Maloney, K.M.; Kalonia, D.S. The effect of Tween® 20 on silicone oil–fusion protein interactions. Int. J. Pharm. 2012, 429, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Dixit, N.; Maloney, K.M.; Kalonia, D.S. Protein-silicone oil interactions: Comparative effect of nonionic surfactants on the interfacial behavior of a fusion protein. Pharm. Res. 2013, 30, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pinnamaneni, S.; Quan, Y.; Jaiswal, A.; Andersson, F.I.; Zhang, X. Mechanistic understanding of protein-silicone oil interactions. Pharm. Res. 2012, 29, 1689–1697. [Google Scholar] [CrossRef]
- Zheng, S.; Puri, A.; Li, J.; Jaiswal, A.; Adams, M. Particle characterization for a protein drug product stored in pre-filled syringes using micro-flow imaging, Archimedes, and quartz crystal microbalance with dissipation. AAPS J. 2016, 19, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Deshmukh, S. Challenges and considerations in development and manufacturing of high concentration biologics drug products. J. Pharm. Innov. 2019, 15, 255–267. [Google Scholar] [CrossRef]
- Patel, A.R.; Kerwin, B.A.; Kanapuram, S.R. Viscoelastic characterization of high concentration antibody formulations using quartz crystal microbalance with dissipation monitoring. J. Pharm. Sci. 2009, 98, 3108–3116. [Google Scholar] [CrossRef]
- Hartl, J.; Peschel, A.; Johannsmann, D.; Garidel, P. Characterizing protein–protein-interaction in high-concentration monoclonal antibody systems with the quartz crystal microbalance. Phys. Chem. Chem. Phys. 2017, 19, 32698–32707. [Google Scholar] [CrossRef] [Green Version]
- Kapp, S.J.; Larsson, I.; Van De Weert, M.; Cárdenas, M.; Jorgensen, L. Competitive adsorption of monoclonal antibodies and nonionic surfactants at solid hydrophobic surfaces. J. Pharm. Sci. 2015, 104, 593–601. [Google Scholar] [CrossRef]
- Rathore, N.; Rajan, R. Current perspectives on stability of protein drug products during formulation, fill and finish operations. Biotechnol. Prog. 2008, 24, 504–514. [Google Scholar] [CrossRef]
- Jameel, F.; Hershenson, S. Formulation and Process Development Strategies for Manufacturing Biopharmaceuticals; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; ISBN 9780470118122. [Google Scholar]
- Oom, A.; Poggi, M.; Wikström, J.; Sukumar, M. Surface interactions of monoclonal antibodies characterized by quartz crystal microbalance with dissipation: Impact of hydrophobicity and protein self-interactions. J. Pharm. Sci. 2012, 101, 519–529. [Google Scholar] [CrossRef]
- Kalonia, C.K.; Heinrich, F.; Curtis, J.E.; Raman, S.; Miller, M.A.; Hudson, S.D. Protein adsorption and layer formation at the stainless steel–solution interface mediates shear-induced particle formation for an IgG1 monoclonal antibody. Mol. Pharm. 2018, 15, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Defante, A.P.; Kalonia, C.K.; Keegan, E.; Bishop, S.M.; Satish, H.A.; Hudson, S.D.; Santacroce, P.V. The impact of the metal interface on the stability and quality of a therapeutic fusion protein. Mol. Pharm. 2020, 17, 569–578. [Google Scholar] [CrossRef] [PubMed]
- QSense Pro. Available online: https://www.biolinscientific.com/qsense/instruments/qsense-pro (accessed on 11 July 2020).
- Lal, G.; Tiwari, D. Investigation of nanoclay doped polymeric composites on piezoelectric Quartz Crystal Microbalance (QCM) sensor. Sens. Actuators B Chem. 2018, 262, 64–69. [Google Scholar] [CrossRef]
- Biolin Scientific, Sensors. Available online: https://www.biolinscientific.com/qsense/sensors (accessed on 11 July 2020).


| Used Techniques | QCM Sensing Layer/System | Aggregating Protein/Peptide | Protein/Peptide Length | Disease/Application | Ref. |
|---|---|---|---|---|---|
| QCM, DLS, AFM | Ure2p covalently bonded to QCM | Ure2p protein, rate of fibril growth | 354 or 359- | Fibril assembly | [78] |
| QCM, AFM, CD | Q-Sense E4, BiolinScientific, Sweden | Amylin aggregates and Au/g-C3N4 | 20–29 | T2DM | [79] |
| QCM | 5-MHz SiO2-coated QCM (Inficon, East Syracuse, NY, USA). | Aβ, binding interactions between SPBs and Aβ proteins | 1–40 | Alzheimer’s disease | [80] |
| QCM | QCM with Aβ1–40 intermediates | Aβ, rate of elongation monitoring | 1–40 | Alzheimer’s disease | [81] |
| QCM, AFM | QCM with short fibril segments | Insulin | 21 + 30 | Injection-localized amyloidosis | [82] |
| QCM-D | In situ multilayer amyloid deposition monitoring | Glucagon | 29 | Regulation of blood, treatment of severe hypoglycemia | [83] |
| TIRE, QCM | DE2 antibodies with PAH | Aβ in the direct immune reaction with monoclonal DE2 antibodies | 1–16 | Alzheimer’s disease | [84] |
| AFM, SPR, QCM-D | Q-Sense E1 BiolinScientific, Sweden | Aβ | 1–42 | Alzheimer’s disease | [85] |
| QCM, Simoa | Silica-coated crystals | Aβ, discrimination between monomers and oligomers | 1–42 | Alzheimer’s disease | [30] |
| QCM, AFM | - | Degradation of Aβ fibrils byphotoactive meso-tetra(4-sulfonatophenyl)porphyrin under UV irradiation | 1–42 | Alzheimer’s disease | [86] |
| QCM-D, Super Resolution Microscopy | QCM immobilized with fibrils | α-synuclein fibrils, secondary nucleation of monomers on fibril surface | 140 | Parkinson’s disease | [87] |
| Application | Method | Instrument | Molecule Type | Molecule Concentration | Ref. |
|---|---|---|---|---|---|
| Evaluation of HPβCD stabilizing properties | QCM-R | QCM200 + QCM25 (Stanford Research Systems) | IgG A, IgG B | 0.1–1.0 mg/mL | [107] |
| Influence of pH and ionic strength on protein-silicone oil interactions | QCM-R | QCM200 (Stanford Research Systems) | Fc-fusion protein | 0.001–1 mg/mL | [112] |
| Influence of polysorbate 20 on protein-silicone oil interactions | QCM-R | QCM200 (Stanford Research Systems) | Fc-fusion protein | 0.1 mg/mL | [113] |
| Influence of polysorbate 20, polysorbate 80 and poloxamer 188 on protein-silicone oil interactions | QCM-R | QCM200 (Stanford Research Systems) | Fc-fusion protein | 0.1 mg/mL | [114] |
| Influence of polysorbate 80 and poloxamer 188 on protein-silicone oil interactions | QCM-D | Q-Sense (Biolin Scientific Inc.) | Abatacept | 1 and 10 mg/mL | [115] |
| Supplementary analysis during particle characterization studies (MFI and Archimedes) of protein drug product stored in prefilled syringes | QCM-D | Q-Sense (Biolin Scientific Inc.) | Therapeutic protein | 0.3 mg/mL | [116] |
| Characterization of viscoelastic properties of HC protein formulation | QCM-D | Q-Sense (Biolin Scientific Inc.) | IgG2 | 70 mg/mL | [118] |
| Characterization of protein-protein interactions in HC protein formulation | QCM-I | QCM sensors (Suzhou SJ Biomaterials Co., Ltd.,) Network analyzer (N2PK design; Makarov Instruments) | Mab1 | 200 mg/mL | [119] |
| Clinical administration material compatibility study | QCM-D | Q-Sense (Biolin Scientific Inc.) | IgG1, IgG2 | 10 mg/mL | [120] |
| Application | Method | Instrument | Molecule Type | Molecule Concentration | Ref. |
|---|---|---|---|---|---|
| Process material compatibility studies | QCM-D | Q-Sense (Biolin Scientific Inc.) | mAb 1, mAb 2 | 1 and 50 mg/mL | [123] |
| QCM-D | Q-Sense (Biolin Scientific Inc.) | NISTmAb | 0.1–100 mg/mL | [124] | |
| QCM-D | Q-Sense (Biolin Scientific Inc.) | Fc-fusion protein | 0.1–110 mg/mL | [125] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Migoń, D.; Wasilewski, T.; Suchy, D. Application of QCM in Peptide and Protein-Based Drug Product Development. Molecules 2020, 25, 3950. https://doi.org/10.3390/molecules25173950
Migoń D, Wasilewski T, Suchy D. Application of QCM in Peptide and Protein-Based Drug Product Development. Molecules. 2020; 25(17):3950. https://doi.org/10.3390/molecules25173950
Chicago/Turabian StyleMigoń, Dorian, Tomasz Wasilewski, and Dariusz Suchy. 2020. "Application of QCM in Peptide and Protein-Based Drug Product Development" Molecules 25, no. 17: 3950. https://doi.org/10.3390/molecules25173950





