A Paradigm for Peptide Hormone-GPCR Analyses
Abstract
:1. Introduction
2. The Mating Pathway in the Yeast Saccharomyces cerevisiae: Relevance to GPCRs
3. Peptide Ligands for Ste2p, the Yeast GPCR for the Peptide Pheromone α-Factor
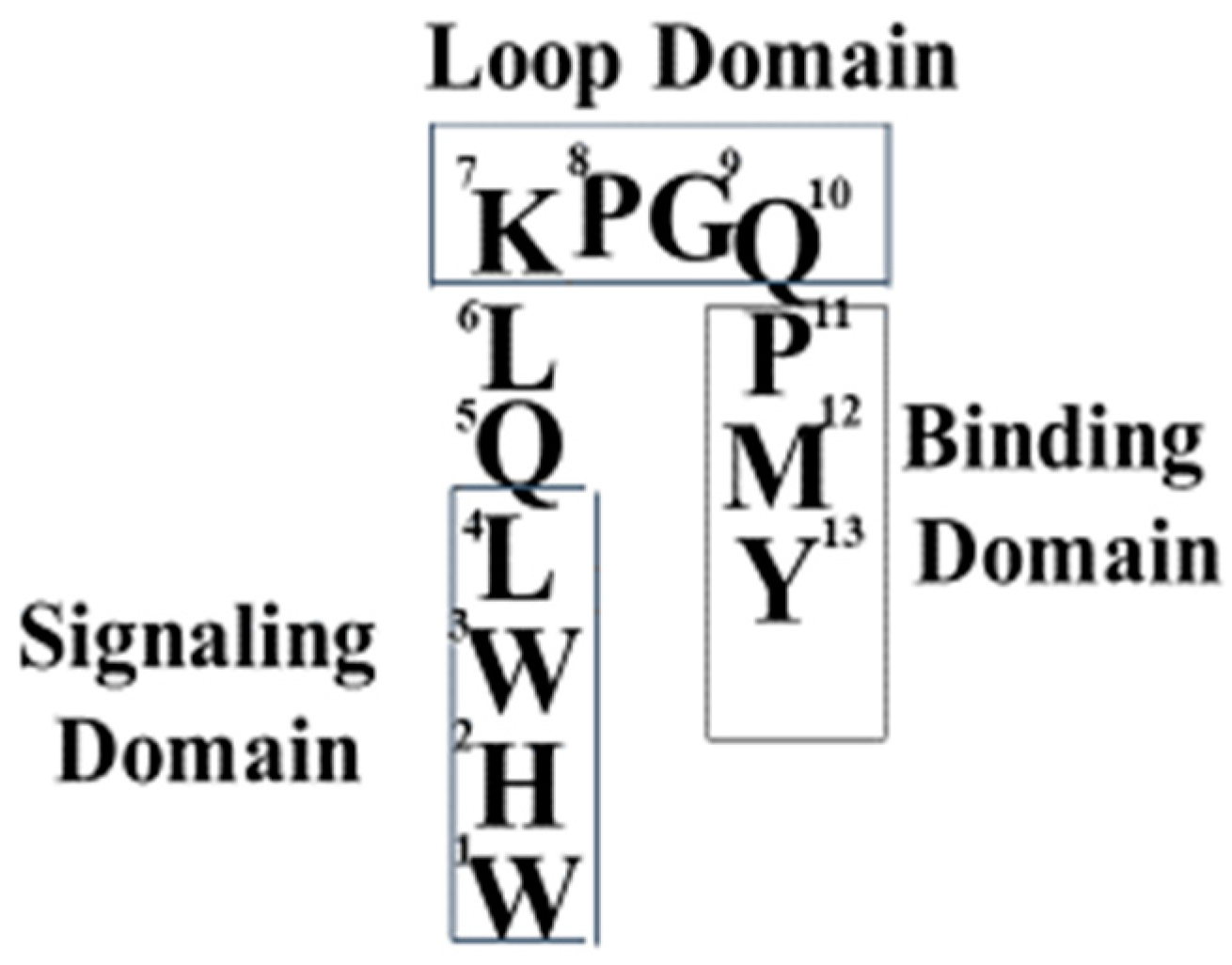
3.1. The Tridecapeptide α-Factor Has Functional Domains
3.2. Conformation of α-Factor at the Step Binding Site
3.3. Discovery of Synergists, Allosteric Ligands, and Biased Agonists
3.3.1. Synergists/Allosteric Ligands
3.3.2. Biased Agonists
4. Identification of Ligand-Receptor Interaction by Cross-Linking Studies
5. Insights into Ste2p Functional Assemblies from Fluorescence Approaches
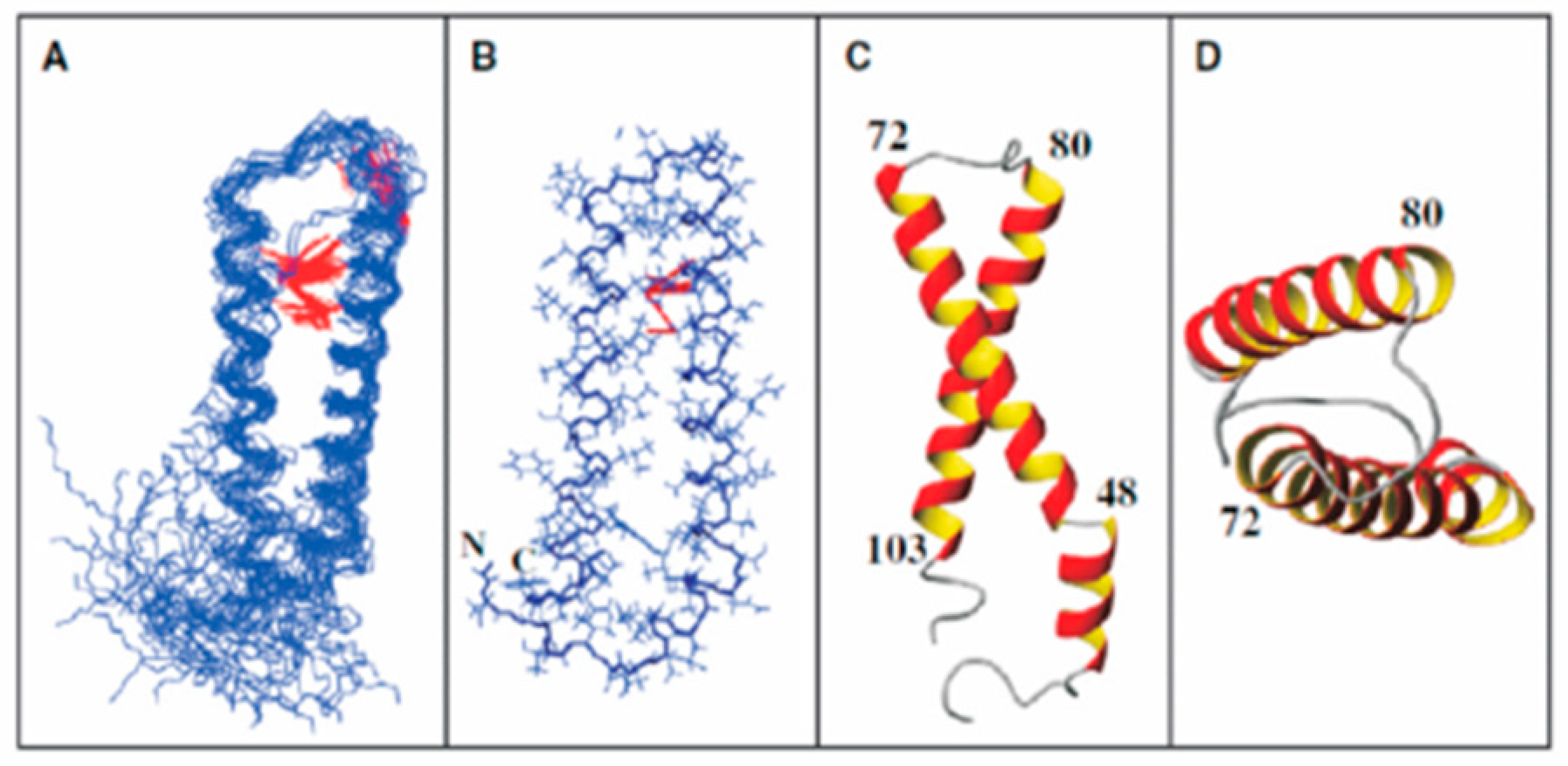
6. Studies of Ste2p Fragments: A Role for Peptide Synthesis and NMR Studies
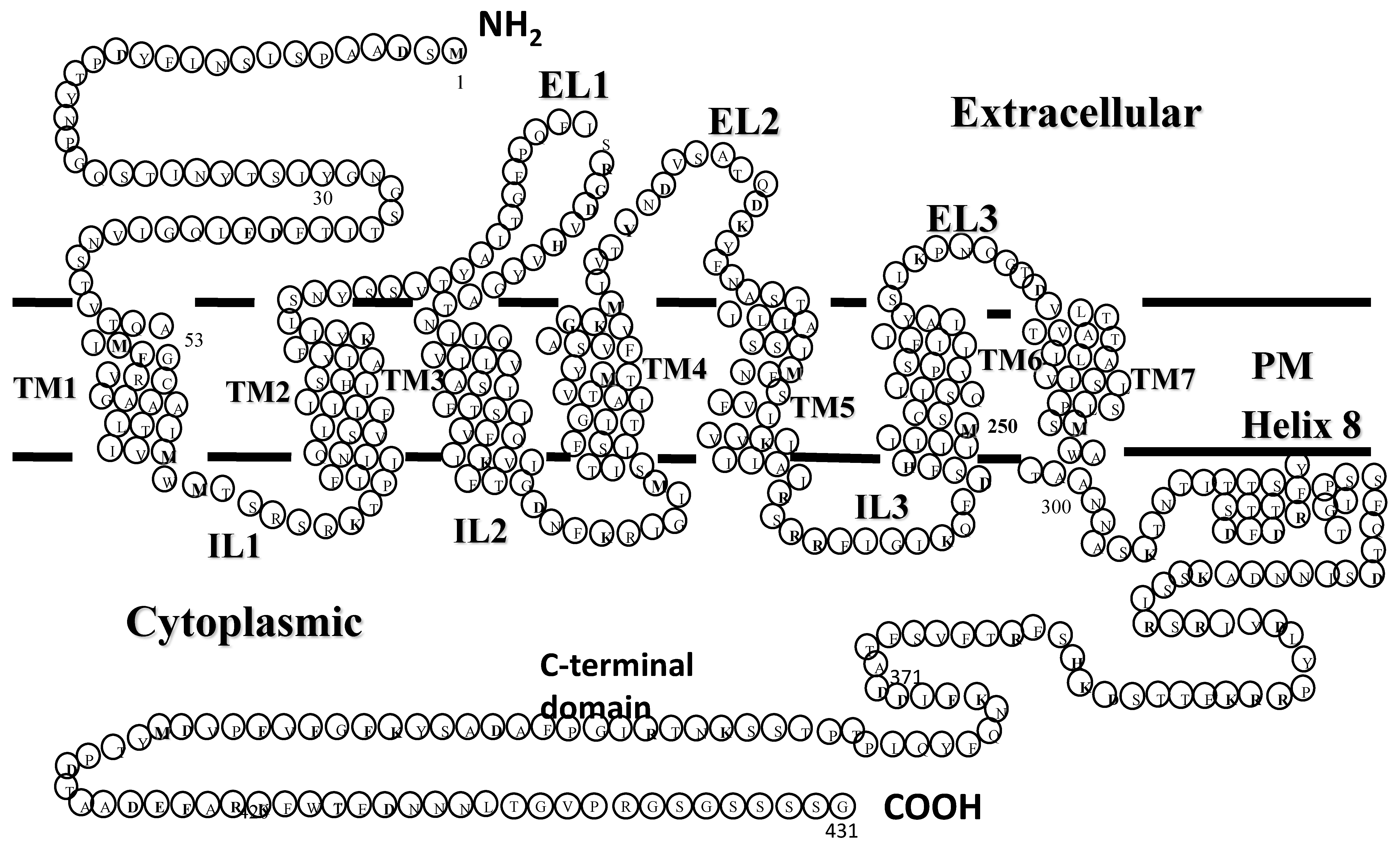
7. Genetic Manipulation of the Receptor Structure
7.1. Identification of Specific Residues Involved in Ligand Binding via Receptor Mutagenesis
7.2. Function and Structure Determination of Ste2p by Mutagenesis
7.2.1. Exploring the Extracellular Region of Ste2p: The N-terminus and EL1
7.2.2. Transmembrane Domains
7.2.3. The C-Terminal Domain
8. Summary and Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Hauser, M.; Narita, V.; Donhardt, A.M.; Naider, F.; Becker, J.M. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 2001, 18, 105–112. [Google Scholar] [CrossRef]
- Alvaro, C.G.; Thorner, J. Heterotrimeric G Protein-coupled Receptor Signaling in Yeast Mating Pheromone Response. J. Biol. Chem. 2016, 291, 7788–7795. [Google Scholar] [CrossRef] [Green Version]
- Bucking-Throm, E.; Duntze, W.; Hartwell, L.H.; Manney, T.R. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp. Cell Res. 1973, 76, 99–110. [Google Scholar] [CrossRef]
- Stotzler, D.; Duntze, W. Isolation and characterization of four related peptides exhibiting alpha factor activity from Saccharomyces cerevisiae. Eur. J. Biochem. 1976, 65, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Masui, Y.; Chino, N.; Sakakibara, S.; Tanaka, T.; Murakami, T.; Kita, H. Synthesis of the mating factor of Saccharomyces cerevisiae and its truncated peptides: The structure-activity relationship. Biochem. Biophys. Res. Commun. 1977, 78, 534–538. [Google Scholar] [CrossRef]
- Masui, Y.; Tanaka, T.; Chino, N.; Kita, H.; Sakakibara, S. Amino acid substitution of mating factor of Saccharomyces cerevisiae structure-activity relationship. Biochem. Biophys. Res. Commun. 1979, 86, 982–987. [Google Scholar] [CrossRef]
- Naider, F.; Becker, J.M. The alpha-factor mating pheromone of Saccharomyces cerevisiae: A model for studying the interaction of peptide hormones and G protein-coupled receptors. Peptides 2004, 25, 1441–1463. [Google Scholar] [CrossRef]
- Hartwell, L.H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 1980, 85, 811–822. [Google Scholar] [CrossRef] [Green Version]
- Jenness, D.D.; Burkholder, A.C.; Hartwell, L.H. Binding of alpha-factor pheromone to yeast a cells: Chemical and genetic evidence for an alpha-factor receptor. Cell 1983, 35, 521–529. [Google Scholar] [CrossRef]
- Jenness, D.D.; Goldman, B.S.; Hartwell, L.H. Saccharomyces cerevisiae mutants unresponsive to alpha-factor pheromone: Alpha-factor binding and extragenic suppression. Mol. Cell Biol. 1987, 7, 1311–1319. [Google Scholar] [CrossRef] [Green Version]
- Blumer, K.J.; Reneke, J.E.; Thorner, J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 10836–10842. [Google Scholar] [PubMed]
- Burkholder, A.C.; Hartwell, L.H. The yeast alpha-factor receptor: Structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985, 13, 8463–8475. [Google Scholar] [CrossRef]
- Nakayama, N.; Miyajima, A.; Arai, K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985, 4, 2643–2648. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Kobilka, B.K.; Strader, D.J.; Benovic, J.L.; Dohlman, H.G.; Frielle, T.; Bolanowski, M.A.; Bennett, C.D.; Rands, E.; Diehl, R.E.; et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature 1986, 321, 75–79. [Google Scholar] [CrossRef]
- Liu, R.; Wong, W.; AP, I.J. Human G protein-coupled receptor studies in Saccharomyces cerevisiae. Biochem. Pharm. 2016, 114, 103–115. [Google Scholar] [CrossRef]
- Jeansonne, N.E. Yeast as a model system for mammalian seven-transmembrane segment receptors. Proc. Soc. Exp. Biol. Med. 1994, 206, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, H.G.; Thorner, J.; Caron, M.G.; Lefkowitz, R.J. Model systems for the study of seven-transmembrane-segment receptors. Annu. Rev. Biochem. 1991, 60, 653–688. [Google Scholar] [CrossRef] [PubMed]
- Julius, D.; Blair, L.; Brake, A.; Sprague, G.; Thorner, J. Yeast alpha factor is processed from a larger precursor polypeptide: The essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 1983, 32, 839–852. [Google Scholar] [CrossRef]
- Abel, M.G.; Zhang, Y.L.; Lu, H.F.; Naider, F.; Becker, J.M. Structure-function analysis of the Saccharomyces cerevisiae tridecapeptide pheromone using alanine-scanned analogs. J. Pept. Res. 1998, 52, 95–106. [Google Scholar] [CrossRef]
- Naider, F.; Becker, J.M.; Lee, Y.H.; Horovitz, A. Double-mutant cycle scanning of the interaction of a peptide ligand and its G protein-coupled receptor. Biochemistry 2007, 46, 3476–3481. [Google Scholar] [CrossRef] [Green Version]
- Blanpain, C.; Doranz, B.J.; Bondue, A.; Govaerts, C.; De Leener, A.; Vassart, G.; Doms, R.W.; Proudfoot, A.; Parmentier, M. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 2003, 278, 5179–5187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, R.; Nasr, N.; Hadden, D.; Sinfield, J.; Abidi, F.; Al-Sabah, S.; de Maturana, R.L.; Treece-Birch, J.; Willshaw, A.; Donnelly, D. Peptide binding at the GLP-1 receptor. Biochem. Soc. Trans. 2007, 35, 713–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coopman, K.; Wallis, R.; Robb, G.; Brown, A.J.; Wilkinson, G.F.; Timms, D.; Willars, G.B. Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: Modelling the ligand-bound receptor. Mol. Endocrinol. 2011, 25, 1804–1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loumaye, E.; Thorner, J.; Catt, K.J. Yeast mating pheromone activates mammalian gonadotrophs: Evolutionary conservation of a reproductive hormone? Science 1982, 218, 1323–1325. [Google Scholar] [CrossRef] [PubMed]
- Higahijima, T.; Fujimura, K.; Masui, Y.; Sakakibara, S.; Miyazawa, T. Physiological activities of peptides are correlated with the conformations of membrane-bound molecules. FEBS Lett. 1983, 159, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Shenbagamurthi, P.; Kundu, B.; Raths, S.; Becker, J.M.; Naider, F. Biological activity and conformational isomerism in position 9 analogues of the des-1-tryptophan,3-beta-cyclohexylalanine-alpha-factor from Saccharomyces cerevisiae. Biochemistry 1985, 24, 7070–7076. [Google Scholar] [CrossRef]
- Naider, F.; Jelicks, L.A.; Becker, J.M.; Broido, M.S. Biologically significant conformation of the Saccharomyces cerevisiae alpha-factor. Biopolymers 1989, 28, 487–497. [Google Scholar] [CrossRef]
- Jelicks, L.A.; Broido, M.S.; Becker, J.M.; Naider, F.R. Interaction of the Saccharomyces cerevisiae alpha-factor with phospholipid vesicles as revealed by proton and phosphorus NMR. Biochemistry 1989, 28, 4233–4240. [Google Scholar] [CrossRef]
- Gounarides, J.S.; Broido, M.S.; Becker, J.M.; Naider, F.R. Conformational analysis of [d-Ala9]alpha-factor and [l-Ala9]alpha-factor in solution and in the presence of lipid. Biochemistry 1993, 32, 908–917. [Google Scholar] [CrossRef]
- Wakamatsu, K.; Okada, A.; Suzuki, M.; Higashijima, T.; Masui, Y.; Sakakibara, S.; Miyazawa, T. Nuclear-magnetic-resonance studies on the conformation of membrane-bound alpha-mating factor. Transferred nuclear Overhauser effect analysis. Eur. J. Biochem. 1986, 154, 607–615. [Google Scholar] [CrossRef]
- Wakamatsu, K.; Okada, A.; Miyazawa, T.; Masui, Y.; Sakakibara, S.; Higashijima, T. Conformations of yeast alpha-mating factor and analog peptides as bound to phospholipid bilayer. Correlation of membrane-bound conformation with physiological activity. Eur. J. Biochem. 1987, 163, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Higashijima, T.; Masui, Y.; Chino, N.; Sakakibara, S.; Kita, H.; Miyazawa, T. Nuclear-magnetic-resonance studies on the conformations of tridecapeptide alpha-mating factor from yeast Saccharomyces cerevisiae and analog peptides in aqueous solution. Conformation-activity relationship. Eur. J. Biochem. 1984, 140, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Schwyzer, R. In search of the ‘bio-active conformation’—Is it induced by the target cell membrane? J. Mol. Recognit 1995, 8, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Bader, R.; Zerbe, O. Are hormones from the neuropeptide Y family recognized by their receptors from the membrane-bound state? Chembiochem 2005, 6, 1520–1534. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.B.; Eriotou-Bargiota, E.; Miller, D.; Becker, J.M.; Naider, F. A covalently constrained congener of the Saccharomyces cerevisiae tridecapeptide mating pheromone is an agonist. J. Biol. Chem. 1989, 264, 19161–19168. [Google Scholar]
- Yang, W.; McKinney, A.; Becker, J.M.; Naider, F. Systematic analysis of the Saccharomyces cerevisiae alpha-factor containing lactam constraints of different ring size. Biochemistry 1995, 34, 1308–1315. [Google Scholar] [CrossRef]
- Eriotou-Bargiota, E.; Xue, C.B.; Naider, F.; Becker, J.M. Antagonistic and synergistic peptide analogues of the tridecapeptide mating pheromone of Saccharomyces cerevisiae. Biochemistry 1992, 31, 551–557. [Google Scholar] [CrossRef]
- Thal, D.M.; Glukhova, A.; Sexton, P.M.; Christopoulos, A. Structural insights into G-protein-coupled receptor allostery. Nature 2018, 559, 45–53. [Google Scholar] [CrossRef]
- Pocklington, M.J.; Orr, E. Novobiocin activates the mating response in yeast through the alpha-pheromone receptor, Ste2p. Biochim. Biophys. Acta 1994, 1224, 401–412. [Google Scholar] [CrossRef]
- Lin, J.C.; Parrish, W.; Eilers, M.; Smith, S.O.; Konopka, J.B. Aromatic residues at the extracellular ends of transmembrane domains 5 and 6 promote ligand activation of the G protein-coupled alpha-factor receptor. Biochemistry 2003, 42, 293–301. [Google Scholar] [CrossRef]
- Rymer, J.K.; Hauser, M.; Bourdon, A.K.; Campagna, S.R.; Naider, F.; Becker, J.M. Novobiocin and peptide analogs of alpha-factor are positive allosteric modulators of the yeast G protein-coupled receptor Ste2p. Biochim. Biophys. Acta 2015, 1848, 916–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisler, J.W.; Xiao, K.; Thomsen, A.R.; Lefkowitz, R.J. Recent developments in biased agonism. Curr. Opin. Cell Biol. 2014, 27, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, G.; Levy, F.O. Novel aspects of G-protein-coupled receptor signalling–different ways to achieve specificity. Acta Physiol. 2007, 190, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Baffi, R.A.; Shenbagamurthi, P.; Terrance, K.; Becker, J.M.; Naider, F.; Lipke, P.N. Different structure-function relationships for alpha-factor-induced morphogenesis and agglutination in Saccharomyces cerevisiae. J. Bacteriol. 1984, 158, 1152–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvaro, C.G.; O’Donnell, A.F.; Prosser, D.C.; Augustine, A.A.; Goldman, A.; Brodsky, J.L.; Cyert, M.S.; Wendland, B.; Thorner, J. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell Biol. 2014, 34, 2660–2681. [Google Scholar] [CrossRef] [Green Version]
- Kiefer, H.; Lindstrom, J.; Lennox, E.S.; Singer, S.J. Photo-affinity labeling of specific acetylcholine-binding sites on membranes. Proc. Natl. Acad. Sci. USA 1970, 67, 1688–1694. [Google Scholar] [CrossRef] [Green Version]
- Singer, S.J.; Doolittle, R.F. Antibody active sites and immunoglobulin molecules. Science 1966, 153, 13–25. [Google Scholar] [CrossRef]
- Singh, A.; Thornton, E.R.; Westheimer, F.H. The photolysis of diazoacetylchymotrypsin. J. Biol. Chem. 1962, 237, 3006–3008. [Google Scholar]
- Chowdhry, V.; Vaughan, R.; Westheimer, F.H. 2-diazo-3,3,3-trifluoropropionyl chloride: Reagent for photoaffinity labeling. Proc. Natl. Acad. Sci. USA 1976, 73, 1406–1408. [Google Scholar] [CrossRef] [Green Version]
- Pattison, D.I.; Davies, M.J. Actions of ultraviolet light on cellular structures. In Cancer: Cell Structures, Carcinogens and Genomic Instability; Birkhäuser: Basel, Switzerland, 2006; pp. 131–157. [Google Scholar]
- Dorman, G.; Nakamura, H.; Pulsipher, A.; Prestwich, G.D. The Life of Pi Star: Exploring the Exciting and Forbidden Worlds of the Benzophenone Photophore. Chem. Rev. 2016, 116, 15284–15398. [Google Scholar] [CrossRef]
- Henry, L.K.; Khare, S.; Son, C.; Babu, V.V.; Naider, F.; Becker, J.M. Identification of a contact region between the tridecapeptide alpha-factor mating pheromone of Saccharomyces cerevisiae and its G protein-coupled receptor by photoaffinity labeling. Biochemistry 2002, 41, 6128–6139. [Google Scholar] [CrossRef] [PubMed]
- Naider, F.; Becker, J.M. Structure-activity relationships of the yeast alpha-factor. Crc. Crit. Rev. Biochem. 1986, 21, 225–248. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Henry, L.K.; Lee, B.K.; Wang, S.H.; Arshava, B.; Becker, J.M.; Naider, F. Position 13 analogs of the tridecapeptide mating pheromone from Saccharomyces cerevisiae: Design of an iodinatable ligand for receptor binding. J. Pept. Res. 2000, 56, 24–34. [Google Scholar] [CrossRef]
- Son, C.D.; Sargsyan, H.; Naider, F.; Becker, J.M. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry 2004, 43, 13193–13203. [Google Scholar] [CrossRef] [PubMed]
- Burdine, L.; Gillette, T.G.; Lin, H.J.; Kodadek, T. Periodate-triggered cross-linking of DOPA-containing peptide-protein complexes. J. Am. Chem. Soc. 2004, 126, 11442–11443. [Google Scholar] [CrossRef]
- Lim, H.S.; Cai, D.; Archer, C.T.; Kodadek, T. Periodate-triggered cross-linking reveals Sug2/Rpt4 as the molecular target of a peptoid inhibitor of the 19S proteasome regulatory particle. J. Am. Chem. Soc. 2007, 129, 12936–12937. [Google Scholar] [CrossRef] [Green Version]
- Umanah, G.K.; Son, C.; Ding, F.; Naider, F.; Becker, J.M. Cross-linking of a DOPA-containing peptide ligand into its G protein-coupled receptor. Biochemistry 2009, 48, 2033–2044. [Google Scholar] [CrossRef] [Green Version]
- Umanah, G.K.; Huang, L.; Ding, F.X.; Arshava, B.; Farley, A.R.; Link, A.J.; Naider, F.; Becker, J.M. Identification of residue-to-residue contact between a peptide ligand and its G protein-coupled receptor using periodate-mediated dihydroxyphenylalanine cross-linking and mass spectrometry. J. Biol. Chem. 2010, 285, 39425–39436. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Shin, Y.O.; Hanson, J.; Cass, B.; Loewen, M.C.; Durocher, Y. Purification and characterization of a recombinant G-protein-coupled receptor, Saccharomyces cerevisiae Ste2p, transiently expressed in HEK293 EBNA1 cells. Biochemistry 2005, 44, 15705–15714. [Google Scholar] [CrossRef]
- Ding, F.X.; Lee, B.K.; Hauser, M.; Davenport, L.; Becker, J.M.; Naider, F. Probing the binding domain of the Saccharomyces cerevisiae alpha-mating factor receptor with rluorescent ligands. Biochemistry 2001, 40, 1102–1108. [Google Scholar] [CrossRef]
- Ding, F.X.; Lee, B.K.; Hauser, M.; Patri, R.; Arshava, B.; Becker, J.M.; Naider, F. Study of the binding environment of alpha-factor in its G protein-coupled receptor using fluorescence spectroscopy. J. Pept. Res. 2002, 60, 65–74. [Google Scholar] [CrossRef]
- Kiriakidi, S.; Kolocouris, A.; Liapakis, G.; Ikram, S.; Durdagi, S.; Mavromoustakos, T. Effects of Cholesterol on GPCR Function: Insights from Computational and Experimental Studies. Adv. Exp. Med. Biol. 2019, 1135, 89–103. [Google Scholar]
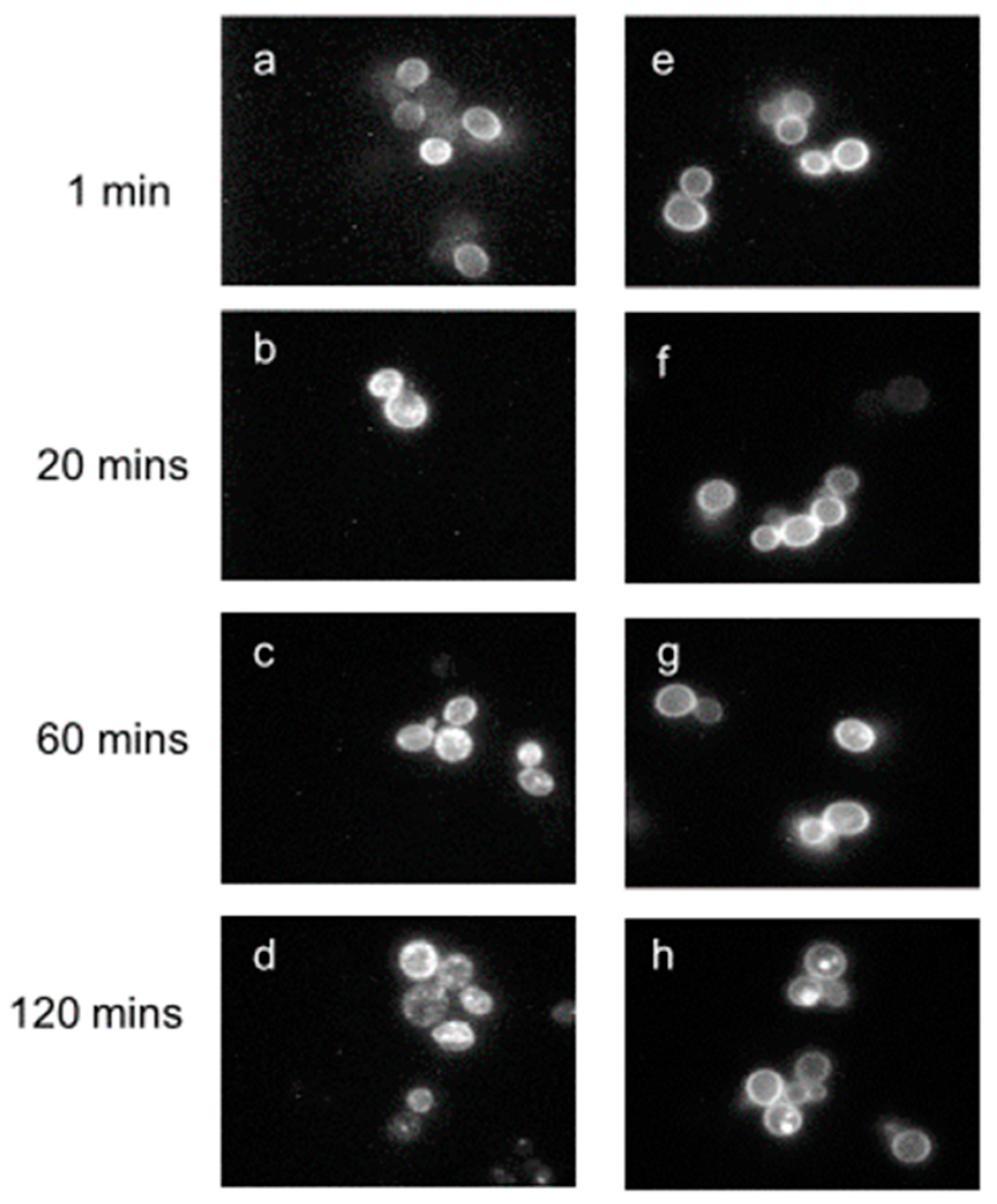
- Schandel, K.A.; Jenness, D.D. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol. Cell Biol. 1994, 14, 7245–7255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yesilaltay, A.; Jenness, D.D. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol. Biol. Cell 2000, 11, 2873–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geli, M.I.; Riezman, H. Endocytic internalization in yeast and animal cells: Similar and different. J. Cell Sci. 1998, 111, 1031–1037. [Google Scholar] [PubMed]
- Jenness, D.D.; Spatrick, P. Down regulation of the alpha-factor pheromone receptor in S. cerevisiae. Cell 1986, 46, 345–353. [Google Scholar] [CrossRef]
- Raths, S.K.; Naider, F.; Becker, J.M. Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 17333–17341. [Google Scholar]
- Jenness, D.D.; Burkholder, A.C.; Hartwell, L.H. Binding of alpha-factor pheromone to Saccharomyces cerevisiae a cells: Dissociation constant and number of binding sites. Mol. Cell Biol. 1986, 6, 318–320. [Google Scholar] [CrossRef]
- Bajaj, A.; Celic, A.; Ding, F.X.; Naider, F.; Becker, J.M.; Dumont, M.E. A fluorescent alpha-factor analogue exhibits multiple steps on binding to its G protein coupled receptor in yeast. Biochemistry 2004, 43, 13564–13578. [Google Scholar] [CrossRef]
- Mathew, E.; Bajaj, A.; Connelly, S.M.; Sargsyan, H.; Ding, F.X.; Hajduczok, A.G.; Naider, F.; Dumont, M.E. Differential interactions of fluorescent agonists and antagonists with the yeast G protein coupled receptor Ste2p. J. Mol. Biol. 2011, 409, 513–528. [Google Scholar] [CrossRef] [Green Version]
- Henderson, R.; Baldwin, J.M.; Ceska, T.A.; Zemlin, F.; Beckmann, E.; Downing, K.H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990, 213, 899–929. [Google Scholar] [CrossRef]
- Ceska, T.A.; Henderson, R.; Baldwin, J.M.; Zemlin, F.; Beckmann, E.; Downing, K. An atomic model for the structure of bacteriorhodopsin, a seven-helix membrane protein. Acta Physiol. Scand. Suppl. 1992, 607, 31–40. [Google Scholar] [PubMed]
- Grigorieff, N.; Ceska, T.A.; Downing, K.H.; Baldwin, J.M.; Henderson, R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 1996, 259, 393–421. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.L.; Gerchman, S.E.; Engelman, D.M. Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J. Mol. Biol. 1987, 198, 655–676. [Google Scholar] [CrossRef]
- Zen, K.H.; McKenna, E.; Bibi, E.; Hardy, D.; Kaback, H.R. Expression of lactose permease in contiguous fragments as a probe for membrane-spanning domains. Biochemistry 1994, 33, 8198–8206. [Google Scholar] [CrossRef]
- Kobilka, B.K.; Kobilka, T.S.; Daniel, K.; Regan, J.W.; Caron, M.G.; Lefkowitz, R.J. Chimeric alpha 2-,beta 2-adrenergic receptors: Delineation of domains involved in effector coupling and ligand binding specificity. Science 1988, 240, 1310–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridge, K.D.; Lee, S.S.; Yao, L.L. In vivo assembly of rhodopsin from expressed polypeptide fragments. Proc. Natl. Acad. Sci. USA 1995, 92, 3204–3208. [Google Scholar] [CrossRef] [Green Version]
- Martin, N.P.; Leavitt, L.M.; Sommers, C.M.; Dumont, M.E. Assembly of G protein-coupled receptors from fragments: Identification of functional receptors with discontinuities in each of the loops connecting transmembrane segments. Biochemistry 1999, 38, 682–695. [Google Scholar] [CrossRef]
- Reddy, A.P.; Tallon, M.A.; Becker, J.M.; Naider, F. Biophysical studies on fragments of the alpha-factor receptor protein. Biopolymers 1994, 34, 679–689. [Google Scholar] [CrossRef]
- Cohen, L.S.; Fracchiolla, K.E.; Becker, J.; Naider, F. Invited review: GPCR structural characterization: Using fragments as building blocks to determine a complete structure. Biopolymers 2014, 102, 223–243. [Google Scholar] [CrossRef]
- Cohen, L.S.; Becker, J.M.; Naider, F. Biosynthesis of peptide fragments of eukaryotic GPCRs in Escherichia coli by directing expression into inclusion bodies. J. Pept. Sci. 2010, 16, 213–218. [Google Scholar] [PubMed]
- Naider, F.; Arshava, B.; Ding, F.X.; Arevalo, E.; Becker, J.M. Peptide fragments as models to study the structure of a G-protein coupled receptor: The alpha-factor receptor of Saccharomyces cerevisiae. Biopolymers 2001, 60, 334–350. [Google Scholar] [CrossRef]
- Arshava, B.; Liu, S.F.; Jiang, H.; Breslav, M.; Becker, J.M.; Naider, F. Structure of segments of a G protein-coupled receptor: CD and NMR analysis of the Saccharomyces cerevisiae tridecapeptide pheromone receptor. Biopolymers 1998, 46, 343–357. [Google Scholar] [CrossRef]
- Arshava, B.; Taran, I.; Xie, H.; Becker, J.M.; Naider, F. High resolution NMR analysis of the seven transmembrane domains of a heptahelical receptor in organic-aqueous medium. Biopolymers 2002, 64, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Naider, F.; Khare, S.; Arshava, B.; Severino, B.; Russo, J.; Becker, J.M. Synthetic peptides as probes for conformational preferences of domains of membrane receptors. Biopolymers 2005, 80, 199–213. [Google Scholar] [CrossRef]
- Ding, F.X.; Xie, H.; Arshava, B.; Becker, J.M.; Naider, F. ATR-FTIR study of the structure and orientation of transmembrane domains of the Saccharomyces cerevisiae alpha-mating factor receptor in phospholipids. Biochemistry 2001, 40, 8945–8954. [Google Scholar] [CrossRef]
- Hunt, J.F.; Earnest, T.N.; Bousche, O.; Kalghatgi, K.; Reilly, K.; Horvath, C.; Rothschild, K.J.; Engelman, D.M. A biophysical study of integral membrane protein folding. Biochemistry 1997, 36, 15156–15176. [Google Scholar] [CrossRef]
- Yeagle, P.L.; Albert, A.D. G-protein coupled receptor structure. Biochim. Biophys. Acta 2007, 1768, 808–824. [Google Scholar] [CrossRef] [Green Version]
- Valiyaveetil, F.I.; MacKinnon, R.; Muir, T.W. Semisynthesis and folding of the potassium channel KcsA. J. Am. Chem. Soc. 2002, 124, 9113–9120. [Google Scholar] [CrossRef]
- Zheng, J.S.; Yu, M.; Qi, Y.K.; Tang, S.; Shen, F.; Wang, Z.P.; Xiao, L.; Zhang, L.; Tian, C.L.; Liu, L. Expedient total synthesis of small to medium-sized membrane proteins via Fmoc chemistry. J. Am. Chem. Soc. 2014, 136, 3695–3704. [Google Scholar] [CrossRef]
- Zuo, C.; Tang, S.; Zheng, J.S. Chemical synthesis and biophysical applications of membrane proteins. J. Pept. Sci. 2015, 21, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Staley, J.P.; Kim, P.S. Formation of a native-like subdomain in a partially folded intermediate of bovine pancreatic trypsin inhibitor. Protein Sci. 1994, 3, 1822–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Marassi, F.M.; Jones, D.H.; Straus, S.K.; Bour, S.; Strebel, K.; Schubert, U.; Oblatt-Montal, M.; Montal, M.; Opella, S.J. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002, 11, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Fracchiolla, K.E.; Cohen, L.S.; Arshava, B.; Poms, M.; Zerbe, O.; Becker, J.M.; Naider, F. Structural characterization of triple transmembrane domain containing fragments of a yeast G protein-coupled receptor in an organic: Aqueous environment by solution-state NMR spectroscopy. J. Pept. Sci. 2015, 21, 212–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocherla, H.; Marino, J.; Shao, X.; Graf, J.; Zou, C.; Zerbe, O. Biosynthesis and spectroscopic characterization of 2-TM fragments encompassing the sequence of a human GPCR, the Y4 receptor. Chembiochem 2012, 13, 818–828. [Google Scholar] [CrossRef] [PubMed]
- Englander, J.; Cohen, L.; Arshava, B.; Estephan, R.; Becker, J.M.; Naider, F. Selective labeling of a membrane peptide with 15N-amino acids using cells grown in rich medium. Biopolymers 2006, 84, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, R.A.; Partridge, A.W.; Yip, J.; Wu, Y.; Goto, N.K.; Deber, C.M. Polar residue tagging of transmembrane peptides. Biopolymers 2003, 71, 675–685. [Google Scholar] [CrossRef]
- de Planque, M.R.; Goormaghtigh, E.; Greathouse, D.V.; Koeppe, R.E., 2nd; Kruijtzer, J.A.; Liskamp, R.M.; de Kruijff, B.; Killian, J.A. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry 2001, 40, 5000–5010. [Google Scholar] [CrossRef] [Green Version]
- Ingwall, R.T.; Scheraga, H.A.; Lotan, N.; Berger, A.; Katchalski, E. Conformational studies of poly-l-alanine in water. Biopolymers 1968, 6, 331–368. [Google Scholar] [CrossRef]
- Neumoin, A.; Arshava, B.; Becker, J.; Zerbe, O.; Naider, F. NMR studies in dodecylphosphocholine of a fragment containing the seventh transmembrane helix of a G-protein-coupled receptor from Saccharomyces cerevisiae. Biophys. J. 2007, 93, 467–482. [Google Scholar] [CrossRef] [Green Version]
- Palczewski, K.; Kumasaka, T.; Hori, T.; Behnke, C.A.; Motoshima, H.; Fox, B.A.; Le Trong, I.; Teller, D.C.; Okada, T.; Stenkamp, R.E.; et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 2000, 289, 739–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, A.G.; Menon, S.T.; Terry, T.J.; Sakmar, T.P. Evidence that helix 8 of rhodopsin acts as a membrane-dependent conformational switch. Biochemistry 2002, 41, 8298–8309. [Google Scholar] [CrossRef]
- Luttrell, L.M.; Lefkowitz, R.J. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 2002, 115, 455–465. [Google Scholar] [PubMed]
- Hicke, L.; Zanolari, B.; Riezman, H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 1998, 141, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, V.V.; Gurevich, E.V. GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front. Pharm. 2019, 10, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.M.; Lee, Y.H.; Akal-Strader, A.; Uddin, M.S.; Hauser, M.; Naider, F.; Becker, J.M. Multiple regulatory roles of the carboxy terminus of Ste2p a yeast GPCR. Pharm. Res. 2012, 65, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Overton, M.C.; Chinault, S.L.; Blumer, K.J. Oligomerization, biogenesis, and signaling is promoted by a glycophorin A-like dimerization motif in transmembrane domain 1 of a yeast G protein-coupled receptor. J. Biol. Chem. 2003, 278, 49369–49377. [Google Scholar] [CrossRef] [Green Version]
- Teese, M.G.; Langosch, D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry 2015, 54, 5125–5135. [Google Scholar] [CrossRef]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein-Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Audet, M.; Stevens, R.C. Emerging structural biology of lipid G protein-coupled receptors. Protein Sci. 2019, 28, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Opella, S.J.; Marassi, F.M. Applications of NMR to membrane proteins. Arch. Biochem. Biophys. 2017, 628, 92–101. [Google Scholar] [CrossRef]
- Goodman, M.; Verdini, A.S.; Toniolo, C.; Phillips, W.D.; Bovey, F.A. Sensitive criteria for the critical size for helix formation in oligopeptides. Proc. Natl. Acad. Sci. USA 1969, 64, 444–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estephan, R.; Englander, J.; Arshava, B.; Samples, K.L.; Becker, J.M.; Naider, F. Biosynthesis and NMR analysis of a 73-residue domain of a Saccharomyces cerevisiae G protein-coupled receptor. Biochemistry 2005, 44, 11795–11810. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, P.A.; Opella, S.J. Effect of detergent concentration on multidimensional solution NMR spectra of membrane proteins in micelles. J. Magn. Reson. Ser. B 1993, 102, 120–125. [Google Scholar] [CrossRef]
- Page, R.C.; Moore, J.D.; Nguyen, H.B.; Sharma, M.; Chase, R.; Gao, F.P.; Mobley, C.K.; Sanders, C.R.; Ma, L.; Sonnichsen, F.D.; et al. Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J. Struct. Funct. Genom. 2006, 7, 51–64. [Google Scholar] [CrossRef]
- Vinogradova, O.; Sonnichsen, F.; Sanders, C.R. 2nd On choosing a detergent for solution NMR studies of membrane proteins. J. Biomol. Nmr 1998, 11, 381–386. [Google Scholar] [CrossRef]
- Killian, J.A.; Trouard, T.P.; Greathouse, D.V.; Chupin, V.; Lindblom, G. A general method for the preparation of mixed micelles of hydrophobic peptides and sodium dodecyl sulphate. FEBS Lett. 1994, 348, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Poms, M.; Ansorge, P.; Martinez-Gil, L.; Jurt, S.; Gottstein, D.; Fracchiolla, K.E.; Cohen, L.S.; Guntert, P.; Mingarro, I.; Naider, F.; et al. NMR Investigation of Structures of G-protein Coupled Receptor Folding Intermediates. J. Biol. Chem. 2016, 291, 27170–27186. [Google Scholar] [CrossRef] [Green Version]
- Cohen, L.S.; Arshava, B.; Neumoin, A.; Becker, J.M.; Guntert, P.; Zerbe, O.; Naider, F. Comparative NMR analysis of an 80-residue G protein-coupled receptor fragment in two membrane mimetic environments. Biochim. Biophys. Acta 2011, 1808, 2674–2684. [Google Scholar] [CrossRef] [Green Version]
- Neumoin, A.; Cohen, L.S.; Arshava, B.; Tantry, S.; Becker, J.M.; Zerbe, O.; Naider, F. Structure of a double transmembrane fragment of a G-protein-coupled receptor in micelles. Biophys. J. 2009, 96, 3187–3196. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Naider, F.; Zerbe, O. Biosynthesis and NMR-studies of a double transmembrane domain from the Y4 receptor, a human GPCR. J. Biomol. Nmr 2008, 42, 257–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, X.; Zou, C.; Naider, F.; Zerbe, O. Comparison of fragments comprising the first two helices of the human Y4 and the yeast Ste2p G-protein-coupled receptors. Biophys. J. 2012, 103, 817–826. [Google Scholar] [CrossRef] [Green Version]
- Marino, J.; Walser, R.; Poms, M.; Zerbe, O. Understanding GPCR Recognition and Folding from NMR Studies of Fragments. RSC Adv. 2018, 8, 9858–9870. [Google Scholar] [CrossRef]
- Whiteway, M.; Hougan, L.; Dignard, D.; Bell, L.; Saari, G.; Grant, F.; O’Hara, P.; MacKay, V.L.; Thomas, D.Y. Function of the STE4 and STE18 genes in mating pheromone signal transduction in Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 1988, 53, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Blumer, K.J.; Thorner, J. Beta and gamma subunits of a yeast guanine nucleotide-binding protein are not essential for membrane association of the alpha subunit but are required for receptor coupling. Proc. Natl. Acad. Sci. USA 1990, 87, 4363–4367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konopka, J.B.; Jenness, D.D. Genetic fine-structural analysis of the Saccharomyces cerevisiae alpha-pheromone receptor. Cell Regul. 1991, 2, 439–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommers, C.M.; Dumont, M.E. Genetic interactions among the transmembrane segments of the G protein coupled receptor encoded by the yeast STE2 gene. J. Mol. Biol. 1997, 266, 559–575. [Google Scholar] [CrossRef]
- Konopka, J.B.; Margarit, S.M.; Dube, P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc. Natl. Acad. Sci. USA 1996, 93, 6764–6769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, L. Substitutions in the hydrophobic core of the alpha-factor receptor of Saccharomyces cerevisiae permit response to Saccharomyces kluyveri alpha-factor and to antagonist. Mol. Cell Biol. 1992, 12, 3959–3966. [Google Scholar] [CrossRef] [Green Version]
- Sen, M.; Shah, A.; Marsh, L. Two types of alpha-factor receptor determinants for pheromone specificity in the mating-incompatible yeasts S. cerevisiae and S. kluyveri. Curr. Genet. 1997, 31, 235–240. [Google Scholar] [CrossRef]
- Sen, M.; Marsh, L. Noncontiguous domains of the alpha-factor receptor of yeasts confer ligand specificity. J. Biol. Chem. 1994, 269, 968–973. [Google Scholar] [PubMed]
- Sridharan, R.; Connelly, S.M.; Naider, F.; Dumont, M.E. Variable Dependence of Signaling Output on Agonist Occupancy of Ste2p, a G Protein-coupled Receptor in Yeast. J. Biol. Chem. 2016, 291, 24261–24279. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.K.; Lee, Y.H.; Hauser, M.; Son, C.D.; Khare, S.; Naider, F.; Becker, J.M. Tyr266 in the sixth transmembrane domain of the yeast alpha-factor receptor plays key roles in receptor activation and ligand specificity. Biochemistry 2002, 41, 13681–13689. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.K.; Khare, S.; Naider, F.; Becker, J.M. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to alpha-factor pheromone binding. J. Biol. Chem. 2001, 276, 37950–37961. [Google Scholar] [PubMed]
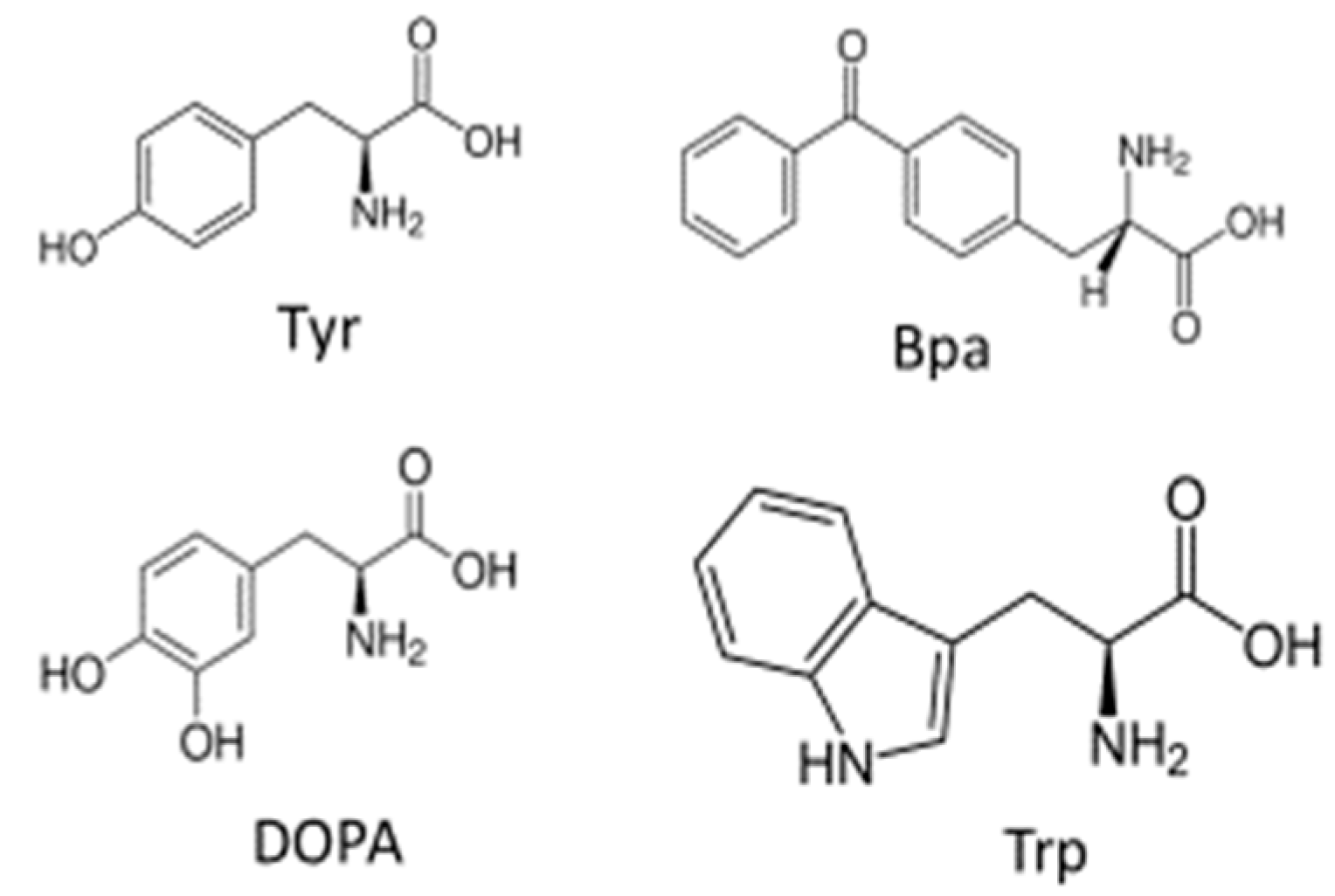
- Huang, L.Y.; Umanah, G.; Hauser, M.; Son, C.; Arshava, B.; Naider, F.; Becker, J.M. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry 2008, 47, 5638–5648. [Google Scholar] [CrossRef]
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [Green Version]
- Koole, C.; Reynolds, C.A.; Mobarec, J.C.; Hick, C.; Sexton, P.M.; Sakmar, T.P. Genetically encoded photocross-linkers determine the biological binding site of exendin-4 peptide in the N-terminal domain of the intact human glucagon-like peptide-1 receptor (GLP-1R). J. Biol. Chem. 2017, 292, 7131–7144. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, L.; Cao, Y.; Cho, A.; Sedki, D.; Huber, T.; Sakmar, T.P.; Laporte, S.A. Genetic code expansion and photocross-linking identify different beta-arrestin binding modes to the angiotensin II type 1 receptor. J. Biol. Chem. 2019, 294, 17409–17420. [Google Scholar] [CrossRef]
- Daggett, K.A.; Sakmar, T.P. Site-specific in vitro and in vivo incorporation of molecular probes to study G-protein-coupled receptors. Curr. Opin. Chem. Biol. 2011, 15, 392–398. [Google Scholar] [CrossRef]
- Grunbeck, A.; Sakmar, T.P. Probing G protein-coupled receptor-ligand interactions with targeted photoactivatable cross-linkers. Biochemistry 2013, 52, 8625–8632. [Google Scholar] [CrossRef] [PubMed]
- Zuber, J.; Danial, S.A.; Connelly, S.M.; Naider, F.; Dumont, M.E. Identification of destabilizing and stabilizing mutations of Ste2p, a G protein-coupled receptor in Saccharomyces cerevisiae. Biochemistry 2015, 54, 1787–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, P.; Kozlovskii, I.; Katritch, V. Computational design for thermostabilization of GPCRs. Curr. Opin. Struct. Biol. 2019, 55, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Magnani, F.; Serrano-Vega, M.J.; Shibata, Y.; Abdul-Hussein, S.; Lebon, G.; Miller-Gallacher, J.; Singhal, A.; Strege, A.; Thomas, J.A.; Tate, C.G. A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat. Protoc. 2016, 11, 1554–1571. [Google Scholar] [CrossRef]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta 2007, 1768, 794–807. [Google Scholar] [CrossRef] [Green Version]
- Lagerstrom, M.C.; Schioth, H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008, 7, 339–357. [Google Scholar] [CrossRef]
- Coleman, J.L.; Ngo, T.; Smith, N.J. The G protein-coupled receptor N-terminus and receptor signalling: N-tering a new era. Cell Signal. 2017, 33, 1–9. [Google Scholar] [CrossRef]
- Shi, C.; Kaminskyj, S.; Caldwell, S.; Loewen, M.C. A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proc. Natl. Acad. Sci. USA 2007, 104, 5395–5400. [Google Scholar] [CrossRef] [Green Version]
- Mentesana, P.E.; Konopka, J.B. Mutational analysis of the role of N-glycosylation in alpha-factor receptor function. Biochemistry 2001, 40, 9685–9694. [Google Scholar] [CrossRef]
- Cartwright, C.P.; Tipper, D.J. In vivo topological analysis of Ste2, a yeast plasma membrane protein, by using beta-lactamase gene fusions. Mol. Cell Biol. 1991, 11, 2620–2628. [Google Scholar] [CrossRef]
- Uddin, M.S.; Hauser, M.; Naider, F.; Becker, J.M. The N-terminus of the yeast G protein-coupled receptor Ste2p plays critical roles in surface expression, signaling, and negative regulation. Biochim. Biophys. Acta 2016, 1858, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.S.; Kim, H.; Deyo, A.; Naider, F.; Becker, J.M. Identification of residues involved in homodimer formation located within a beta-strand region of the N-terminus of a Yeast G protein-coupled receptor. J. Recept. Signal. Transduct. Res. 2012, 32, 65–75. [Google Scholar] [CrossRef]
- Akal-Strader, A.; Khare, S.; Xu, D.; Naider, F.; Becker, J.M. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J. Biol. Chem. 2002, 277, 30581–30590. [Google Scholar] [CrossRef] [Green Version]
- Uddin, M.S.; Naider, F.; Becker, J.M. Dynamic roles for the N-terminus of the yeast G protein-coupled receptor Ste2p. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.; Kauffman, S.; Lee, B.K.; Naider, F.; Becker, J.M. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. J. Biol. Chem. 2007, 282, 10387–10397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrish, W.; Eilers, M.; Ying, W.; Konopka, J.B. The cytoplasmic end of transmembrane domain 3 regulates the activity of the Saccharomyces cerevisiae G-protein-coupled alpha-factor receptor. Genetics 2002, 160, 429–443. [Google Scholar]
- Lee, Y.H.; Naider, F.; Becker, J.M. Interacting residues in an activated state of a G protein-coupled receptor. J. Biol. Chem. 2006, 281, 2263–2272. [Google Scholar] [CrossRef] [Green Version]
- Umanah, G.K.; Huang, L.Y.; Maccarone, J.M.; Naider, F.; Becker, J.M. Changes in conformation at the cytoplasmic ends of the fifth and sixth transmembrane helices of a yeast G protein-coupled receptor in response to ligand binding. Biochemistry 2011, 50, 6841–6854. [Google Scholar] [CrossRef] [Green Version]
- Milligan, G.; Ward, R.J.; Marsango, S. GPCR homo-oligomerization. Curr Opin Cell Biol 2019, 57, 40–47. [Google Scholar] [CrossRef]
- Gurevich, V.V.; Gurevich, E.V. GPCRs and Signal Transducers: Interaction Stoichiometry. Trends Pharm. Sci. 2018, 39, 672–684. [Google Scholar] [CrossRef]
- Wang, H.X.; Konopka, J.B. Identification of amino acids at two dimer interface regions of the alpha-factor receptor (Ste2). Biochemistry 2009, 48, 7132–7139. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, B.K.; Naider, F.; Becker, J.M. Identification of specific transmembrane residues and ligand-induced interface changes involved in homo-dimer formation of a yeast G protein-coupled receptor. Biochemistry 2009, 48, 10976–10987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehret, A.U.; Bajaj, A.; Naider, F.; Dumont, M.E. Oligomerization of the yeast alpha-factor receptor: Implications for dominant negative effects of mutant receptors. J. Biol. Chem. 2006, 281, 20698–20714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggio, R.; Fasciani, I.; Rossi, M.; Di Gregorio, J.; Pietrantoni, I.; Puca, V.; Flati, V.; Scarselli, M. Variants of G protein-coupled receptors: A reappraisal of their role in receptor regulation. Biochem. Soc. Trans. 2016, 44, 589–594. [Google Scholar] [CrossRef]
- Cevheroglu, O.; Becker, J.M.; Son, C.D. GPCR-Galpha protein precoupling: Interaction between Ste2p, a yeast GPCR, and Gpa1p, its Galpha protein, is formed before ligand binding via the Ste2p C-terminal domain and the Gpa1p N-terminal domain. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2435–2446. [Google Scholar] [CrossRef]
- Ferre, S. The GPCR heterotetramer: Challenging classical pharmacology. Trends Pharm. Sci. 2015, 36, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Konopka, J.B.; Jenness, D.D.; Hartwell, L.H. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell 1988, 54, 609–620. [Google Scholar] [CrossRef]
- Dosil, M.; Schandel, K.A.; Gupta, E.; Jenness, D.D.; Konopka, J.B. The C terminus of the Saccharomyces cerevisiae alpha-factor receptor contributes to the formation of preactivation complexes with its cognate G protein. Mol. Cell Biol. 2000, 20, 5321–5329. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Konopka, J.B. Regulation of the G-protein-coupled alpha-factor pheromone receptor by phosphorylation. Mol. Cell Biol. 1996, 16, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Conner, M.; Hicks, M.R.; Dafforn, T.; Knowles, T.J.; Ludwig, C.; Staddon, S.; Overduin, M.; Gunther, U.L.; Thome, J.; Wheatley, M.; et al. Functional and biophysical analysis of the C-terminus of the CGRP-receptor; a family B GPCR. Biochemistry 2008, 47, 8434–8444. [Google Scholar] [CrossRef]
- Kuwasako, K.; Hay, D.L.; Nagata, S.; Murakami, M.; Kitamura, K.; Kato, J. Functions of third extracellular loop and helix 8 of Family B GPCRs complexed with RAMPs and characteristics of their receptor trafficking. Curr. Protein Pept. Sci. 2013, 14, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kawasaki, T.; Mine, S.; Matsumura, H. Functional Role of the C-Terminal Amphipathic Helix 8 of Olfactory Receptors and Other G Protein-Coupled Receptors. Int. J. Mol. Sci. 2016, 17, 1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovati, G.E.; Capra, V.; Neubig, R.R. The highly conserved DRY motif of class A G protein-coupled receptors: Beyond the ground state. Mol. Pharm. 2007, 71, 959–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilers, M.; Hornak, V.; Smith, S.O.; Konopka, J.B. Comparison of class A and D G protein-coupled receptors: Common features in structure and activation. Biochemistry 2005, 44, 8959–8975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robles, L.M.; Millán-Pacheco, C.; Pastor, N.; Del Río, G. Structure-Function Studies of the Alpha Pheromone Receptor from Yeast. Tip Rev. Espec. En Cienc. Quim.-Biol. 2017, 20, 16–26. [Google Scholar]
- Ziarek, J.J.; Kleist, A.B.; London, N.; Raveh, B.; Montpas, N.; Bonneterre, J.; St-Onge, G.; DiCosmo-Ponticello, C.J.; Koplinski, C.A.; Roy, I.; et al. Structural basis for chemokine recognition by a G protein-coupled receptor and implications for receptor activation. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Abayev, M.; Rodrigues, J.; Srivastava, G.; Arshava, B.; Jaremko, L.; Jaremko, M.; Naider, F.; Levitt, M.; Anglister, J. The solution structure of monomeric CCL5 in complex with a doubly sulfated N-terminal segment of CCR5. FEBS J. 2018, 285, 1988–2003. [Google Scholar] [CrossRef] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naider, F.; Becker, J.M. A Paradigm for Peptide Hormone-GPCR Analyses. Molecules 2020, 25, 4272. https://doi.org/10.3390/molecules25184272
Naider F, Becker JM. A Paradigm for Peptide Hormone-GPCR Analyses. Molecules. 2020; 25(18):4272. https://doi.org/10.3390/molecules25184272
Chicago/Turabian StyleNaider, Fred, and Jeffrey M. Becker. 2020. "A Paradigm for Peptide Hormone-GPCR Analyses" Molecules 25, no. 18: 4272. https://doi.org/10.3390/molecules25184272






